DNA sequence
Time Limit : 15000/5000ms (Java/Other) Memory Limit : 32768/32768K (Java/Other)
Total Submission(s) : 15 Accepted Submission(s) : 7
Font: Times New Roman | Verdana | Georgia
Font Size: ← →
Problem Description
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
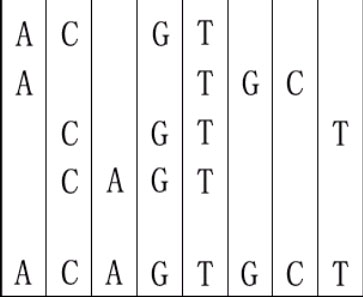
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.

Input
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
Output
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
Sample Input
1 4 ACGT ATGC CGTT CAGT
Sample Output
8
Author
Source
HDU 2006-12 Programming Contest
使用dfs进行搜索,但限制递归深度。
逐步加深搜索深度,直至找到答案。
主函数中, 限制搜索深度,如果无解,就加深1层深度
强力剪枝: 递归函数中, 首先计算最坏情况下,还需要补充长度:
为每个DNA序列还未匹配的长度之和(sum)。
如果现在搜索深度+sum>限定的搜索深度,则返回
#include <iostream> #include<cstdio> #include<cstring> #include<algorithm> using namespace std; char f[4]={'A','T','G','C'}; int flag,i,t,n,maxlen; int cnt[50]; char str[10][10]; void dfs(int len,int cnt[]) { if (flag || len>maxlen) return; int sum=0; for(int i=0;i<n;i++) //关键 :ida*(迭代加深搜索) { int l=strlen(str[i]); sum=max(sum,l-cnt[i]); } if (sum+len>maxlen) return; if (sum==0) {flag=1; return;} for(int i=0;i<4;i++) { char x=f[i]; int next[50]; int tflag=0; for(int j=0;j<n;j++) if (str[j][cnt[j]]==x) { next[j]=cnt[j]+1; tflag=1; } else next[j]=cnt[j]; if (tflag) dfs(len+1,next); //更新了才说明有效 } return; } int main() { scanf("%d",&t); for(;t>0;t--) { scanf("%d",&n); maxlen=0; for(i=0;i<n;i++) { scanf("%s",str[i]); int l=strlen(str[i]); maxlen=max( maxlen,l ); } flag=0; memset(cnt,0,sizeof(cnt)); for(i=0;i<40;i++) { dfs(0,cnt); if (flag) break; maxlen++; } printf("%d\n",maxlen); } return 0; }