1 多项式朴素贝叶斯MultinomialNB
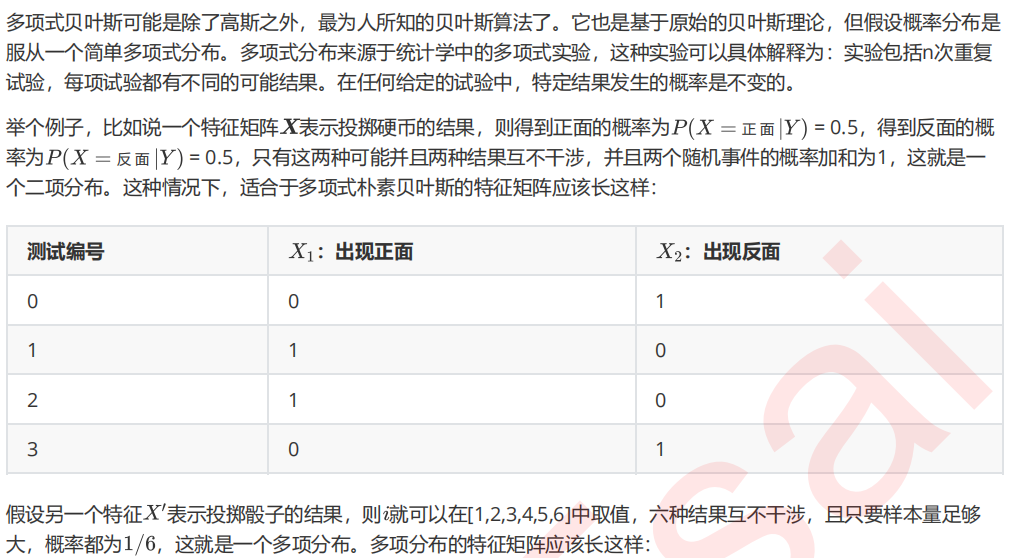


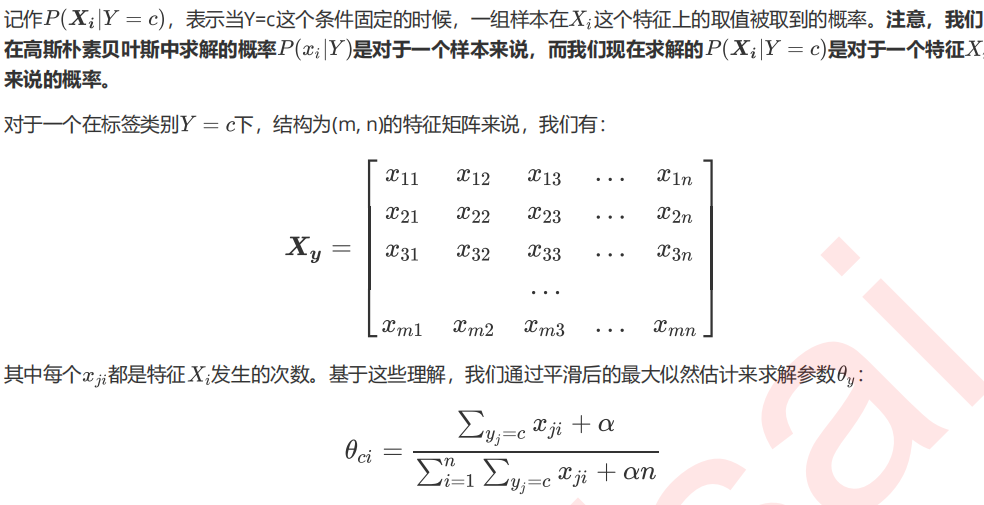


1. 导入需要的模块和库
from sklearn.preprocessing import MinMaxScaler from sklearn.naive_bayes import MultinomialNB from sklearn.model_selection import train_test_split from sklearn.datasets import make_blobs from sklearn.metrics import brier_score_loss
2. 建立数据集
class_1 = 500 class_2 = 500 #两个类别分别设定500个样本 centers = [[0.0, 0.0], [2.0, 2.0]] #设定两个类别的中心 clusters_std = [0.5, 0.5] #设定两个类别的方差 X, y = make_blobs(n_samples=[class_1, class_2], centers=centers, cluster_std=clusters_std, random_state=0, shuffle=False) Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y ,test_size=0.3 ,random_state=420)
3. 归一化,确保输入的矩阵不带有负数
#先归一化,保证输入多项式朴素贝叶斯的特征矩阵中不带有负数 mms = MinMaxScaler().fit(Xtrain) Xtrain_ = mms.transform(Xtrain) Xtest_ = mms.transform(Xtest)
4. 建立一个多项式朴素贝叶斯分类器吧
mnb = MultinomialNB().fit(Xtrain_, Ytrain) #重要属性:调用根据数据获取的,每个标签类的对数先验概率log(P(Y)) #由于概率永远是在[0,1]之间,因此对数先验概率返回的永远是负值 mnb.class_log_prior_ np.unique(Ytrain) (Ytrain == 1).sum()/Ytrain.shape[0] mnb.class_log_prior_.shape #可以使用np.exp来查看真正的概率值 np.exp(mnb.class_log_prior_) #重要属性:返回一个固定标签类别下的每个特征的对数概率log(P(Xi|y)) mnb.feature_log_prob_ mnb.feature_log_prob_.shape #重要属性:在fit时每个标签类别下包含的样本数。当fit接口中的sample_weight被设置时,该接口返回的值也会受 到加权的影响 mnb.class_count_ mnb.class_count_.shape
5. 那分类器的效果如何呢?
#一些传统的接口 mnb.predict(Xtest_) mnb.predict_proba(Xtest_) mnb.score(Xtest_,Ytest) brier_score_loss(Ytest,mnb.predict_proba(Xtest_)[:,1],pos_label=1)
7. 效果不太理想,思考一下多项式贝叶斯的性质,我们能够做点什么呢?
#来试试看把Xtiain转换成分类型数据吧 #注意我们的Xtrain没有经过归一化,因为做哑变量之后自然所有的数据就不会又负数了 from sklearn.preprocessing import KBinsDiscretizer kbs = KBinsDiscretizer(n_bins=10, encode='onehot').fit(Xtrain) Xtrain_ = kbs.transform(Xtrain) Xtest_ = kbs.transform(Xtest) mnb = MultinomialNB().fit(Xtrain_, Ytrain) mnb.score(Xtest_,Ytest) brier_score_loss(Ytest,mnb.predict_proba(Xtest_)[:,1],pos_label=1)

2 伯努利朴素贝叶斯BernoulliNB


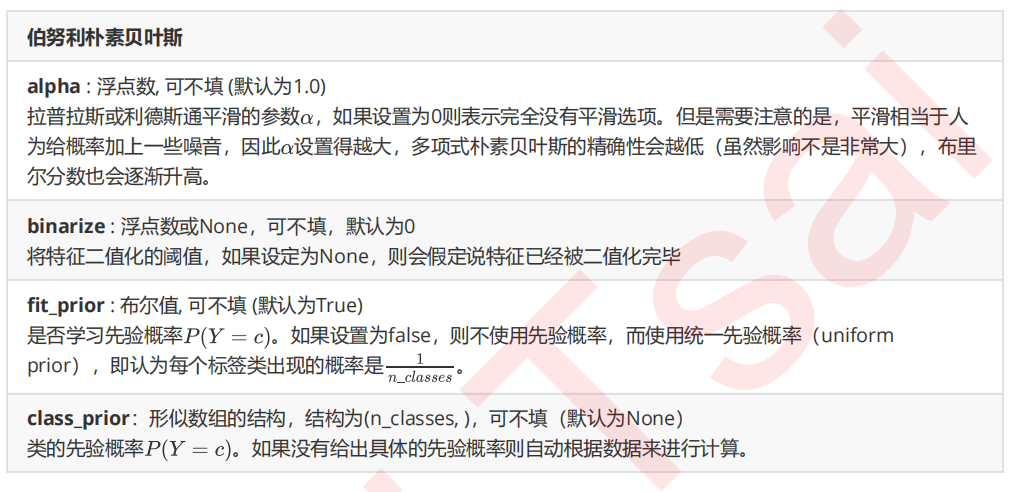
在sklearn中,伯努利朴素贝叶斯的实现也非常简单
from sklearn.naive_bayes import BernoulliNB #普通来说我们应该使用二值化的类sklearn.preprocessing.Binarizer来将特征一个个二值化 #然而这样效率过低,因此我们选择归一化之后直接设置一个阈值 mms = MinMaxScaler().fit(Xtrain) Xtrain_ = mms.transform(Xtrain) Xtest_ = mms.transform(Xtest) #不设置二值化 bnl_ = BernoulliNB().fit(Xtrain_, Ytrain) bnl_.score(Xtest_,Ytest) brier_score_loss(Ytest,bnl_.predict_proba(Xtest_)[:,1],pos_label=1) #设置二值化阈值为0.5 bnl = BernoulliNB(binarize=0.5).fit(Xtrain_, Ytrain) bnl.score(Xtest_,Ytest) brier_score_loss(Ytest,bnl.predict_proba(Xtest_)[:,1],pos_label=1)

3 探索贝叶斯:贝叶斯的样本不均衡问题

1. 导入需要的模块,建立样本不平衡的数据集
from sklearn.naive_bayes import MultinomialNB, GaussianNB, BernoulliNB from sklearn.model_selection import train_test_split from sklearn.datasets import make_blobs from sklearn.preprocessing import KBinsDiscretizer from sklearn.metrics import brier_score_loss as BS,recall_score,roc_auc_score as AUC class_1 = 50000 #多数类为50000个样本 class_2 = 500 #少数类为500个样本 centers = [[0.0, 0.0], [5.0, 5.0]] #设定两个类别的中心 clusters_std = [3, 1] #设定两个类别的方差 X, y = make_blobs(n_samples=[class_1, class_2], centers=centers, cluster_std=clusters_std, random_state=0, shuffle=False) X.shape np.unique(y)
2. 查看所有贝叶斯在样本不平衡数据集上的表现
name = ["Multinomial","Gaussian","Bernoulli"] models = [MultinomialNB(),GaussianNB(),BernoulliNB()] for name,clf in zip(name,models): Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y ,test_size=0.3 ,random_state=420) if name != "Gaussian": kbs = KBinsDiscretizer(n_bins=10, encode='onehot').fit(Xtrain) Xtrain = kbs.transform(Xtrain) Xtest = kbs.transform(Xtest) clf.fit(Xtrain,Ytrain) y_pred = clf.predict(Xtest) proba = clf.predict_proba(Xtest)[:,1] score = clf.score(Xtest,Ytest) print(name) print(" Brier:{:.3f}".format(BS(Ytest,proba,pos_label=1))) print(" Accuracy:{:.3f}".format(score)) print(" Recall:{:.3f}".format(recall_score(Ytest,y_pred))) print(" AUC:{:.3f}".format(AUC(Ytest,proba)))

4 改进多项式朴素贝叶斯:补集朴素贝叶斯ComplementNB


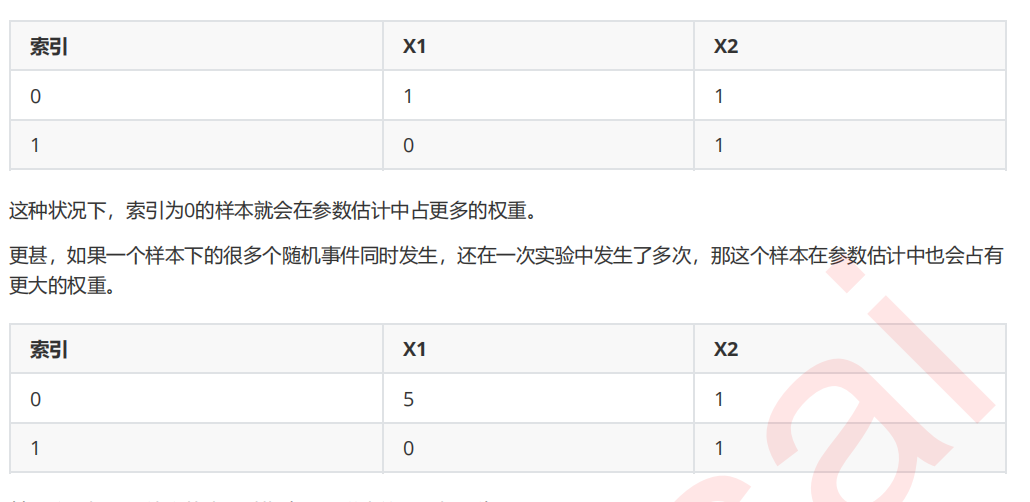

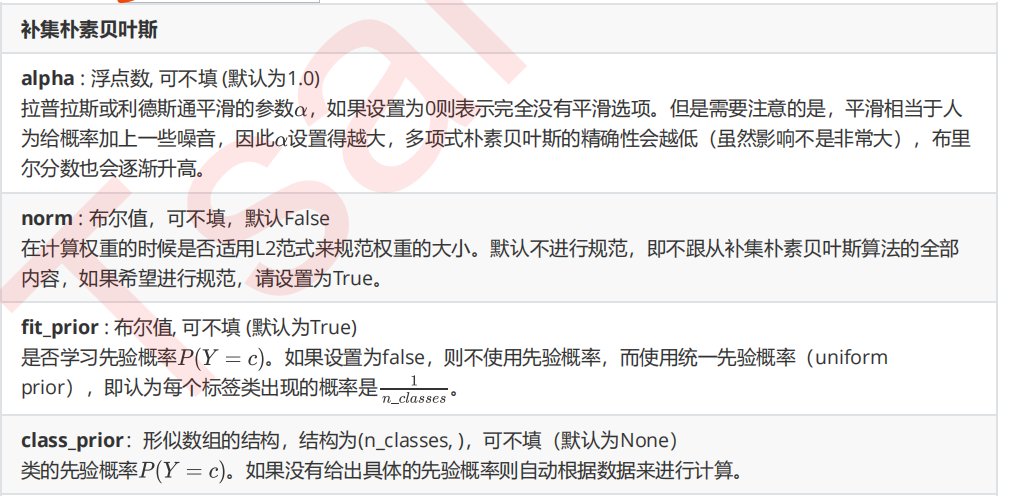

from sklearn.naive_bayes import ComplementNB from time import time import datetime name = ["Multinomial","Gaussian","Bernoulli","Complement"] models = [MultinomialNB(),GaussianNB(),BernoulliNB(),ComplementNB()] for name,clf in zip(name,models): times = time() Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y ,test_size=0.3 ,random_state=420) #预处理 if name != "Gaussian": kbs = KBinsDiscretizer(n_bins=10, encode='onehot').fit(Xtrain) Xtrain = kbs.transform(Xtrain) Xtest = kbs.transform(Xtest) clf.fit(Xtrain,Ytrain) y_pred = clf.predict(Xtest) proba = clf.predict_proba(Xtest)[:,1] score = clf.score(Xtest,Ytest) print(name) print(" Brier:{:.3f}".format(BS(Ytest,proba,pos_label=1))) print(" Accuracy:{:.3f}".format(score)) print(" Recall:{:.3f}".format(recall_score(Ytest,y_pred))) print(" AUC:{:.3f}".format(AUC(Ytest,proba))) print(datetime.datetime.fromtimestamp(time()-times).strftime("%M:%S:%f"))
