Chapter1 WHERE IN THE GENOME DOES DNA REPLICATION BEGIN

一、
·聚合酶启动结构域会结合上游序列的一些位点,这些位点有多个,且特异,并且分布在两条链上。通过计算,找到出现频率最高的k-mer可能为为聚合酶结合位点:dnaA BOX。
但是如何定位Ori的大概位置呢?
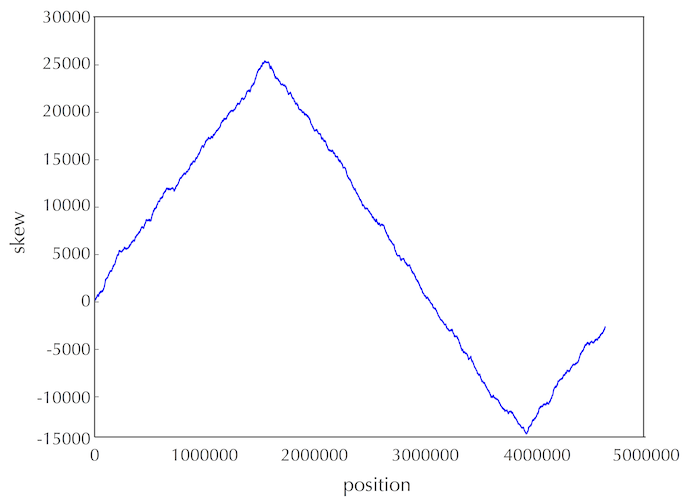
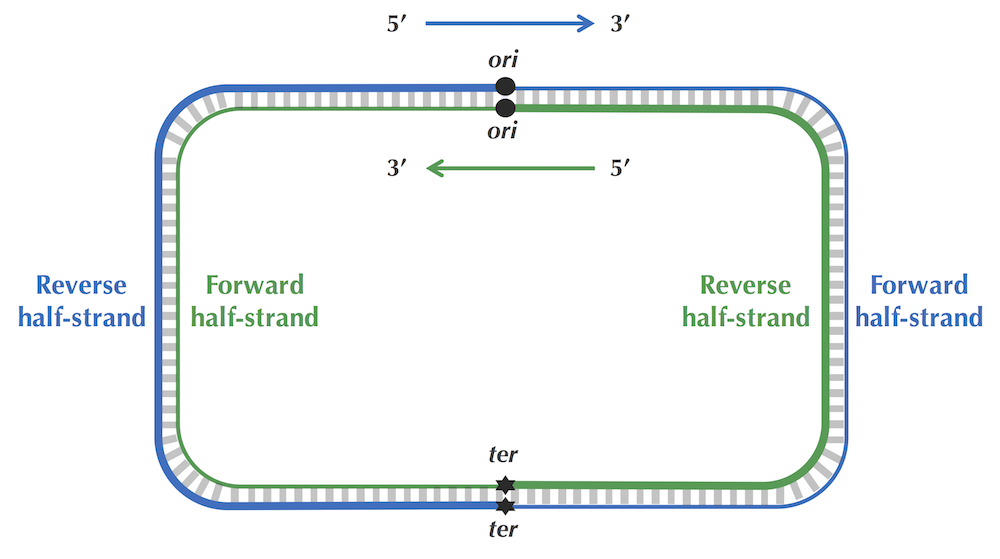
·DNA链复制的不对称性,其导致突变速率的不对称,使得有(forward链C->T,脱氨基)的趋势。由此,依据skew增的处于forward链,skew减的处于reverse链。(skew = G - C ,逢G+1 逢C-1)图中最低点代表Ori区域。
由此可以大致推测出ori的位置,然后在此位置内(100bp),寻找出现频率大的pattern,作为可能的dnaA box。
·由于k-mer之间,会有碱基的若干差异,故应使用能容错的计数方法。
二、
提出问题:The Clump Finding Problem
Find every k-mer that forms a clump in the genome.

ComputingFrequencies(Text, k) #一种遍历一次计算频率的‘桶’方法
for i ← 0 to 4k − 1
FrequencyArray(i) ← 0
for i ← 0 to |Text| − k
Pattern ← Text(i, k)
j ← PatternToNumber(Pattern) #hash
FrequencyArray(j) ← FrequencyArray(j) + 1
return FrequencyArray

FindingFrequentWordsBySorting(Text , k) #排序法
FrequentPatterns ← an empty set
for i ← 0 to |Text| − k
Pattern ← Text(i, k)
Index(i) ← PatternToNumber(Pattern)
Count(i) ← 1
SortedIndex ← Sort(Index)
for i ← 1 to |Text| − k
if SortedIndex(i) = SortedIndex(i − 1)
Count(i) = Count(i − 1) + 1
maxCount ← maximum value in the array Count
for i ← 0 to |Text| − k
if Count(i) = maxCount
Pattern ← NumberToPattern(SortedIndex(i), k)
add Pattern to the set FrequentPatterns
return FrequentPatterns
 一种容错的频率计算方法
一种容错的频率计算方法ClumpFinding(Genome, k, L, t)
FrequentPatterns ← an empty set
for i ← 0 to 4k − 1
Clump(i) ← 0
for i ← 0 to |Genome| − L
Text ← the string of length L starting at position i in Genome
FrequencyArray ← ComputingFrequencies(Text, k)
for index ← 0 to 4k − 1
if FrequencyArray(index) ≥ t
Clump(index) ← 1
for i ← 0 to 4k − 1
if Clump(i) = 1
Pattern ← NumberToPattern(i, k)
add Pattern to the set FrequentPatterns
return FrequentPatterns
我们不用每次挪动一位搜寻窗就重新计算kmer频率,搜寻窗每挪一位,原来的第一个kmer将少一个,结尾后一个kmer将多一个
BetterClumpFinding(Genome, k, t, L)
FrequentPatterns ← an empty set
for i ← 0 to 4k − 1
Clump(i) ← 0
Text ← Genome(0, L)
FrequencyArray ← ComputingFrequencies(Text, k)
for i ← 0 to 4k − 1
if FrequencyArray(i) ≥ t
Clump(i) ← 1
for i ← 1 to |Genome| − L
FirstPattern ← Genome(i − 1, k)
index ← PatternToNumber(FirstPattern)
FrequencyArray(index) ← FrequencyArray(index) − 1
LastPattern ← Genome(i + L − k, k)
index ← PatternToNumber(LastPattern)
FrequencyArray(index) ← FrequencyArray(index) + 1
if FrequencyArray(index) ≥ t
Clump(index) ← 1
for i ← 0 to 4k − 1
if Clump(i) = 1
Pattern ← NumberToPattern(i, k)
add Pattern to the set FrequentPatterns
return FrequentPatterns
