最近比较闲,是时候把自己以前看的资料整理一下了。
LogisticRegression:由于在训练过程中考虑了所有的样本对参数的影响,因此不一定获得最佳的分类器,对比下一篇 svm只用支持向量来帮助决策最优线性分类模型。
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import LogisticRegression
df_train = pd.read_csv('./Breast-Cancer/breast-cancer-train.csv')
df_test = pd.read_csv('./Breast-Cancer/breast-cancer-test.csv')
lr = LogisticRegression()
lr.fit(df_train[['Clump Thickness', 'Cell Size']], df_train['Type'])
print 'Testing accuracy (all training samples):', lr.score(df_test[['Clump Thickness', 'Cell Size']], df_test['Type'])
#使用训练样本学习直线的系数和截距
intercept = lr.intercept_
coef = lr.coef_[0, :]
lx = np.arange(0, 12)
#a*x + b*y = 0
ly = (-intercept - lx * coef[0]) / coef[1]
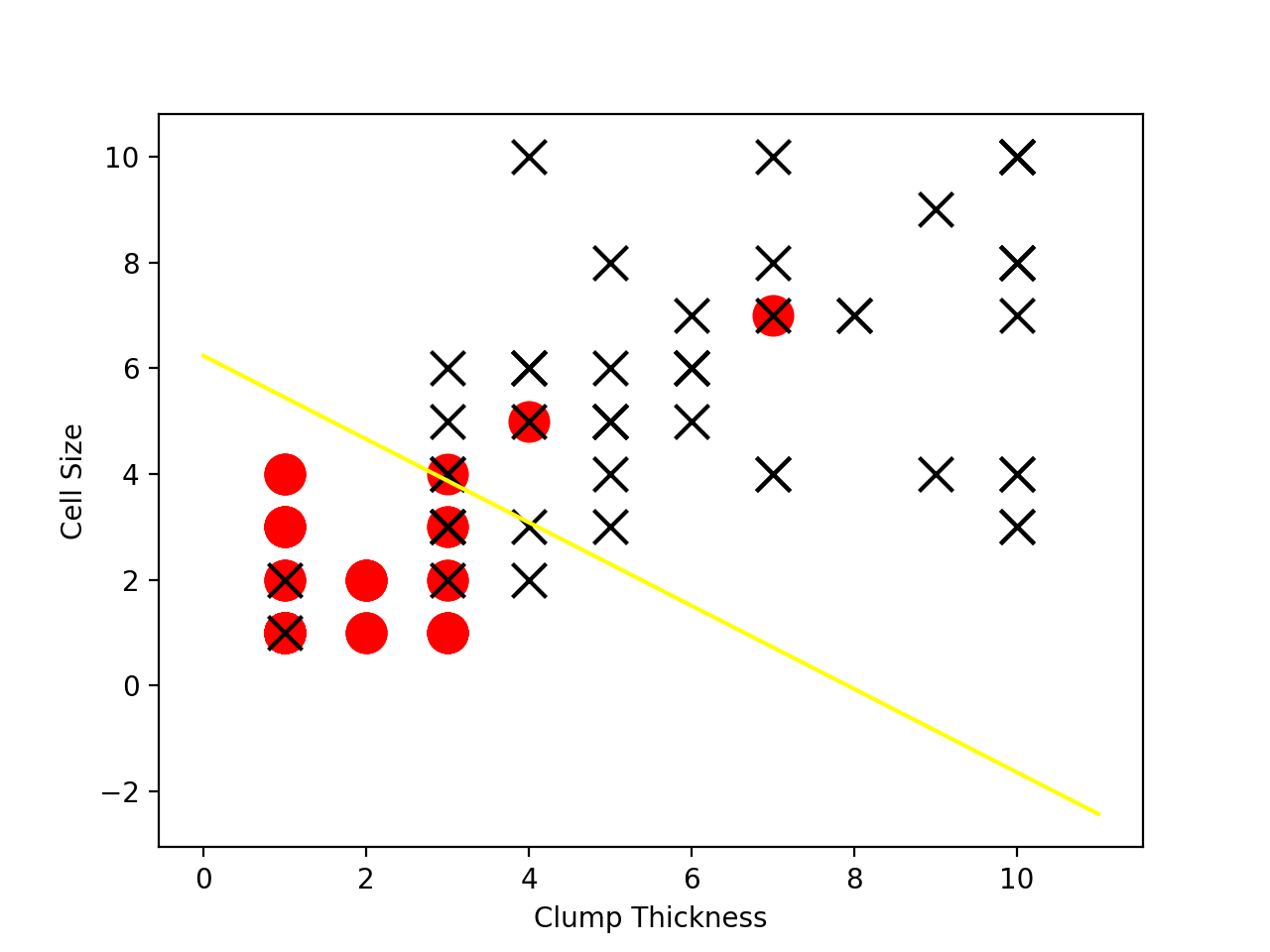
plt.plot(lx, ly, c='yellow')
df_test_negative = df_test.loc[df_test['Type'] == 0][['Clump Thickness', 'Cell Size']]
df_test_positive = df_test.loc[df_test['Type'] == 1][['Clump Thickness', 'Cell Size']]
plt.scatter(df_test_negative['Clump Thickness'], df_test_negative['Cell Size'], marker='o', s=200, c='red')
plt.scatter(df_test_positive['Clump Thickness'], df_test_positive['Cell Size'], marker='x', s=150, c='black')
plt.xlabel('Clump Thickness')
plt.ylabel('Cell Size')
plt.show()
结果如下图: