R语言是单线程的,如果数据量比较大的情况下最好用并行计算来处理数据,这样会获得运行速度倍数的提升。这里介绍一个基于Unix系统的并行程序包:multicore.
我们用三种不同的方式来进行一个简单的数据处理:
我们从 1000 genome project 数据库下载了VCF文件,现在需要手动提取出每个allele的 allele frequency(AF)值(vcftools 可以很好的解决这个问题,但是假设我的vcf文件没有genotype, 或者我要实现一些个性化功能,那么可能要手动解决)。我们仅仅提取VCF文件的前1,000,000行作测试。看一下文件前几行:
##fileformat=VCFv4.1 ##INFO=<ID=LDAF,Number=1,Type=Float,Description="MLE Allele Frequency Accounting for LD"> ##INFO=<ID=AVGPOST,Number=1,Type=Float,Description="Average posterior probability from MaCH/Thunder"> ##INFO=<ID=RSQ,Number=1,Type=Float,Description="Genotype imputation quality from MaCH/Thunder"> ##INFO=<ID=ERATE,Number=1,Type=Float,Description="Per-marker Mutation rate from MaCH/Thunder"> ##INFO=<ID=THETA,Number=1,Type=Float,Description="Per-marker Transition rate from MaCH/Thunder"> ##INFO=<ID=CIEND,Number=2,Type=Integer,Description="Confidence interval around END for imprecise variants"> ##INFO=<ID=CIPOS,Number=2,Type=Integer,Description="Confidence interval around POS for imprecise variants"> ##INFO=<ID=END,Number=1,Type=Integer,Description="End position of the variant described in this record"> ##INFO=<ID=HOMLEN,Number=.,Type=Integer,Description="Length of base pair identical micro-homology at event breakpoints"> ##INFO=<ID=HOMSEQ,Number=.,Type=String,Description="Sequence of base pair identical micro-homology at event breakpoints"> ##INFO=<ID=SVLEN,Number=1,Type=Integer,Description="Difference in length between REF and ALT alleles"> ##INFO=<ID=SVTYPE,Number=1,Type=String,Description="Type of structural variant"> ##INFO=<ID=AC,Number=.,Type=Integer,Description="Alternate Allele Count"> ##INFO=<ID=AN,Number=1,Type=Integer,Description="Total Allele Count"> ##ALT=<ID=DEL,Description="Deletion"> ##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype"> ##FORMAT=<ID=DS,Number=1,Type=Float,Description="Genotype dosage from MaCH/Thunder"> ##FORMAT=<ID=GL,Number=.,Type=Float,Description="Genotype Likelihoods"> ##INFO=<ID=AA,Number=1,Type=String,Description="Ancestral Allele, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/pilot_data/technical/reference/ancestral_alignments/README"> ##INFO=<ID=AF,Number=1,Type=Float,Description="Global Allele Frequency based on AC/AN"> ##INFO=<ID=AMR_AF,Number=1,Type=Float,Description="Allele Frequency for samples from AMR based on AC/AN"> ##INFO=<ID=ASN_AF,Number=1,Type=Float,Description="Allele Frequency for samples from ASN based on AC/AN"> ##INFO=<ID=AFR_AF,Number=1,Type=Float,Description="Allele Frequency for samples from AFR based on AC/AN"> ##INFO=<ID=EUR_AF,Number=1,Type=Float,Description="Allele Frequency for samples from EUR based on AC/AN"> ##INFO=<ID=VT,Number=1,Type=String,Description="indicates what type of variant the line represents"> ##INFO=<ID=SNPSOURCE,Number=.,Type=String,Description="indicates if a snp was called when analysing the low coverage or exome alignment data"> ##reference=GRCh37 #CHROM POS ID REF ALT QUAL FILTER INFO 1 10583 rs58108140 G A 100 PASS AVGPOST=0.7707;RSQ=0.4319;LDAF=0.2327;ERATE=0.0161;AN=2184;VT=SNP;AA=.;THETA=0.0046;AC=314;SNPSOURCE=LOWCOV;AF=0.14;ASN_AF=0.13;AMR_AF=0.17;AFR_AF=0.04;EUR_AF=0.21 1 10611 rs189107123 C G 100 PASS AN=2184;THETA=0.0077;VT=SNP;AA=.;AC=41;ERATE=0.0048;SNPSOURCE=LOWCOV;AVGPOST=0.9330;LDAF=0.0479;RSQ=0.3475;AF=0.02;ASN_AF=0.01;AMR_AF=0.03;AFR_AF=0.01;EUR_AF=0.02 1 13302 rs180734498 C T 100 PASS THETA=0.0048;AN=2184;AC=249;VT=SNP;AA=.;RSQ=0.6281;LDAF=0.1573;SNPSOURCE=LOWCOV;AVGPOST=0.8895;ERATE=0.0058;AF=0.11;ASN_AF=0.02;AMR_AF=0.08;AFR_AF=0.21;EUR_AF=0.14 1 13327 rs144762171 G C 100 PASS AVGPOST=0.9698;AN=2184;VT=SNP;AA=.;RSQ=0.6482;AC=59;SNPSOURCE=LOWCOV;ERATE=0.0012;LDAF=0.0359;THETA=0.0204;AF=0.03;ASN_AF=0.02;AMR_AF=0.03;AFR_AF=0.02;EUR_AF=0.04 1 13957 rs201747181 TC T 28 PASS AA=TC;AC=35;AF=0.02;AFR_AF=0.02;AMR_AF=0.02;AN=2184;ASN_AF=0.01;AVGPOST=0.8711;ERATE=0.0065;EUR_AF=0.02;LDAF=0.0788;RSQ=0.2501;THETA=0.0100;VT=INDEL 1 13980 rs151276478 T C 100 PASS AN=2184;AC=45;ERATE=0.0034;THETA=0.0139;RSQ=0.3603;LDAF=0.0525;VT=SNP;AA=.;AVGPOST=0.9221;SNPSOURCE=LOWCOV;AF=0.02;ASN_AF=0.02;AMR_AF=0.02;AFR_AF=0.01;EUR_AF=0.02 1 30923 rs140337953 G T 100 PASS AC=1584;AA=T;AN=2184;RSQ=0.5481;VT=SNP;THETA=0.0162;SNPSOURCE=LOWCOV;ERATE=0.0183;LDAF=0.6576;AVGPOST=0.7335;AF=0.73;ASN_AF=0.89;AMR_AF=0.80;AFR_AF=0.48;EUR_AF=0.73 1 46402 rs199681827 C CTGT 31 PASS AA=.;AC=8;AF=0.0037;AFR_AF=0.01;AN=2184;ASN_AF=0.0017;AVGPOST=0.8325;ERATE=0.0072;LDAF=0.0903;RSQ=0.0960;THETA=0.0121;VT=INDEL 1 47190 rs200430748 G GA 192 PASS AA=G;AC=29;AF=0.01;AFR_AF=0.06;AMR_AF=0.0028;AN=2184;AVGPOST=0.9041;ERATE=0.0041;LDAF=0.0628;RSQ=0.2883;THETA=0.0153;VT=INDEL
可以发现AF值在INFO这一列字符串里面,所以要取出这个值是比较容易的,只需要对字符串进行切割即可。
先通过data.table包读入数据到计算机内存,我们看到data.table包读入数据非常快,1,000,000行数据读入仅仅5秒。同时函数非常智能的通过后面的格式将前28行识别为注释,没有读入。
library(data.table) head_vcf <- fread("head_1000000.vcf", sep = " ", colClasses=list(character= 1,6))
### Read 1000000 rows and 8 (of 8) columns from 0.166 GB file in 00:00:05
head(head_vcf)
#CHROM POS ID REF ALT QUAL FILTER
1: 1 10583 rs58108140 G A 100 PASS
2: 1 10611 rs189107123 C G 100 PASS
3: 1 13302 rs180734498 C T 100 PASS
4: 1 13327 rs144762171 G C 100 PASS
5: 1 13957 rs201747181 TC T 28 PASS
6: 1 13980 rs151276478 T C 100 PASS
INFO
1: AVGPOST=0.7707;RSQ=0.4319;LDAF=0.2327;ERATE=0.0161;AN=2184;VT=SNP;AA=.;THETA=0.0046;AC=314;SNPSOURCE=LOWCOV;AF=0.14;ASN_AF=0.13;AMR_AF=0.17;AFR_AF=0.04;EUR_AF=0.21
2: AN=2184;THETA=0.0077;VT=SNP;AA=.;AC=41;ERATE=0.0048;SNPSOURCE=LOWCOV;AVGPOST=0.9330;LDAF=0.0479;RSQ=0.3475;AF=0.02;ASN_AF=0.01;AMR_AF=0.03;AFR_AF=0.01;EUR_AF=0.02
3: THETA=0.0048;AN=2184;AC=249;VT=SNP;AA=.;RSQ=0.6281;LDAF=0.1573;SNPSOURCE=LOWCOV;AVGPOST=0.8895;ERATE=0.0058;AF=0.11;ASN_AF=0.02;AMR_AF=0.08;AFR_AF=0.21;EUR_AF=0.14
4: AVGPOST=0.9698;AN=2184;VT=SNP;AA=.;RSQ=0.6482;AC=59;SNPSOURCE=LOWCOV;ERATE=0.0012;LDAF=0.0359;THETA=0.0204;AF=0.03;ASN_AF=0.02;AMR_AF=0.03;AFR_AF=0.02;EUR_AF=0.04
5: AA=TC;AC=35;AF=0.02;AFR_AF=0.02;AMR_AF=0.02;AN=2184;ASN_AF=0.01;AVGPOST=0.8711;ERATE=0.0065;EUR_AF=0.02;LDAF=0.0788;RSQ=0.2501;THETA=0.0100;VT=INDEL
6: AN=2184;AC=45;ERATE=0.0034;THETA=0.0139;RSQ=0.3603;LDAF=0.0525;VT=SNP;AA=.;AVGPOST=0.9221;SNPSOURCE=LOWCOV;AF=0.02;ASN_AF=0.02;AMR_AF=0.02;AFR_AF=0.01;EUR_AF=0.02
第一种方法是使用内建函数,如果使用单核处理数据,那么这是最推荐的方式,内建函数都是用底层语言写好封装并优化的,运算速度非常快。
#编写一个函数,函数的参数info_str是读入文件的第八列(INFO),这是一个字符串向量
get_af_fun1 <- function(info_str) { split1 <- strsplit(info_str, ";AF=") str1 <- vector(length = length(split1)) for(i in 1:length(str1)) str1[i] <- split1[[i]][2] split2 <- strsplit(str1, ";") str2 <- vector(length = length(split2)) for(i in 1:length(str2)) str2[i] <- split2[[i]][1] str2 }
system.time(result1 <- get_af_fun1(as.character(head_vcf$INFO)))
用户 系统 流逝 38.944 0.012 38.950
#因为是根据位置索引操作,所以随机抽取三行发现结果一致
> result1[43525] [1] "0.01" > head_vcf[43525,] #CHROM POS ID REF ALT QUAL FILTER 1: 1 3607355 rs3765729 G A 100 PASS INFO 1: AA=G;AN=2184;RSQ=0.7704;VT=SNP;AC=22;LDAF=0.0126;SNPSOURCE=LOWCOV;AVGPOST=0.9929;THETA=0.0020;ERATE=0.0006;AF=0.01;ASN_AF=0.03;AMR_AF=0.02 > head_vcf[654635,] #CHROM POS ID REF ALT QUAL FILTER 1: 1 49595556 rs192800899 G T 100 PASS INFO 1: AA=G;AN=2184;RSQ=0.8739;AC=6;VT=SNP;THETA=0.0006;LDAF=0.0030;SNPSOURCE=LOWCOV;ERATE=0.0003;AVGPOST=0.9992;AF=0.0027;AMR_AF=0.0028;EUR_AF=0.01 > result1[654635] [1] "0.0027"
> head_vcf[9876,] #CHROM POS ID REF ALT QUAL FILTER 1: 1 1322735 rs146506266 G A 100 PASS INFO 1: RSQ=0.9882;AVGPOST=1.0000;THETA=0.0004;SNPSOURCE=LOWCOV,EXOME;AA=G;AN=2184;VT=SNP;LDAF=0.0019;AC=4;ERATE=0.0003;AF=0.0018;ASN_AF=0.01 > result1[9876] [1] "0.0018"

通过第一个例子我们看到,通过向量化运算和内建函数处理一百万行数据仅仅花费40秒不到,而且对于for循环,预先分配内存会极大的提高速度。主要原因是R语言的数据赋值是拷贝而不是引用,因此如果预先对向量分配内存就避免了每次循环都对该向量重新赋值。同时我们看到在程序是单核在计算。
第二个例子是利用R语言内建循环函数lapply(), 算法和第一个例子相似,只不过把for循环用lapply执行。
fun_str1 <- function(str_v) strsplit(str_v, ";AF=")[[1]][2] fun_str2 <- function(str_v) strsplit(str_v, ";")[[1]][1] get_af_fun2 <- function(info_str) { str1 <- unlist(lapply(info_str, fun_str1)) str2 <- unlist(lapply(str1, fun_str2)) str2 } system.time(result2 <- get_af_fun2(as.character(head_vcf$INFO))) 用户 系统 流逝 76.512 0.028 76.556
#随机抽取三行发现结果一致
> result2[543654] [1] "0.0009" > result2[75676] [1] "0.0014" > result2[8765] [1] "0.0018" > head_vcf[543654] #CHROM POS ID REF ALT QUAL FILTER 1: 1 40941743 rs143883355 G A 100 PASS INFO 1: AN=2184;THETA=0.0005;LDAF=0.0010;VT=SNP;AA=.;AVGPOST=0.9998;RSQ=0.9135;SNPSOURCE=LOWCOV;ERATE=0.0003;AC=2;AF=0.0009;AFR_AF=0.0041 > head_vcf[75676] #CHROM POS ID REF ALT QUAL FILTER 1: 1 5569843 rs148207486 C T 100 PASS INFO 1: ERATE=0.0004;RSQ=0.7694;AA=C;AN=2184;VT=SNP;LDAF=0.0019;AVGPOST=0.9989;THETA=0.0012;SNPSOURCE=LOWCOV;AC=3;AF=0.0014;AFR_AF=0.01 > head_vcf[8765] #CHROM POS ID REF ALT QUAL FILTER 1: 1 1254001 rs190286788 G C 100 PASS INFO 1: ERATE=0.0005;RSQ=0.5499;AA=G;AN=2184;AVGPOST=0.9979;VT=SNP;THETA=0.0006;SNPSOURCE=LOWCOV;AC=4;LDAF=0.0017;AF=0.0018;AMR_AF=0.0028;EUR_AF=0.0040

第二个例子我们可以看出,计算这一百万行运行时间大概70多秒,lapply是代替循环的一个非常好的函数, 这里之所以运行速度比第一种方法慢,我们从程序可以看出,是因为对这个长度为一百万的字符串向量分别调用了一百万次fun_str1和fun_str2。程序在函数调用上花费了一定时间,如果将我们自己写的fun_str1, fun_str2函数换成内建函数,速度也会提高不少。
有了第二个例子的铺垫,我们再来介绍第三个例子,也就是利用multicore 包进行并行处理。这个包是基于Unix系统的,这里简单介绍里面一个函数mclapply(), 和lapply()函数用法基本一致。具体看例子:
fun_str1 <- function(str_v) strsplit(str_v, ";AF=")[[1]][2] fun_str2 <- function(str_v) strsplit(str_v, ";")[[1]][1] get_af_fun3 <- function(info_str) { str1 <- unlist(mclapply(info_str, fun_str1, mc.cores = 4)) str2 <- unlist(mclapply(str1, fun_str2, mc.cores = 4)) str2 } system.time(result3 <- get_af_fun3(as.character(head_vcf$INFO)))
用户 系统 流逝 56.816 65.512 39.958
#检查三次运行的结果显示三个结果向量完全一致
> all(result1 == result2)
[1] TRUE
> all(result3 == result2)
[1] TRUE
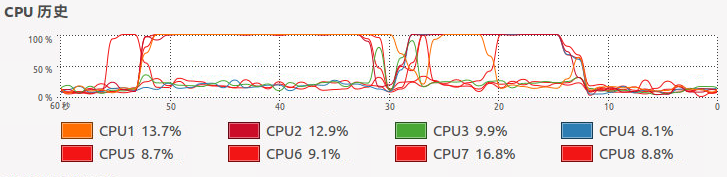
查看CPU使用状态我们可以看到通过mc.cores调用四个核进行并行运算,确实比单核的lapply运算速度提高不少,但是没有四倍的效果是并行运算系统中一些线程活动的额外开销。
再次加大运算量,将文件提高到10,000,000行, 为了避免宕机,这里请用大内存机器进行实验。
head_vcf <- fread("head_10000000.vcf", sep = " ", colClasses=list(character= 1,6))
Read 10000000 rows and 8 (of 8) columns from 1.660 GB file in 00:02:04
方案一,使用内建函数和for循环来处理。
> system.time(result1 <- get_af_fun1(as.character(head_vcf$INFO))) 用户 系统 流逝 603.936 0.248 604.060
方案二,使用lapply()函数单核处理。
> system.time(result2 <- get_af_fun2(as.character(head_vcf$INFO))) 用户 系统 流逝 933.176 0.344 933.115
方案三,使用mclapply()函数并行处理。
> system.time(result3 <- get_af_fun3(as.character(head_vcf$INFO))) 用户 系统 流逝 984.936 251.176 351.119
这个一千万行的例子我们看出,系统内建函数的运行效率还是比lapply快, 开四个核并行的速度最快,而且这个效率还会随着计算核心数的提升而进一步获得较大的提升,有条件的用户可以选择方案三。