期刊:Nature Genetics
影响因子:29.352
一、研究背景
以青蒿素为主的联合疗法一直以来都是治疗疟疾的有效方法,值得关注的是横跨亚洲到非洲都出现了对一线药物的抗药性。阻止出现更高水平的抗药性以及阻止抗药性扩散到非洲刻不容缓。为了更好同抗药性进行斗争,了解遗传因素在抗药性的混合和传播中的作用非常重要。
二、材料
选取来自柬埔寨、越南、老挝、缅甸、孟加拉国、刚果和尼日利亚的13个地方的疟原虫样本1063(取自血液如下图)。 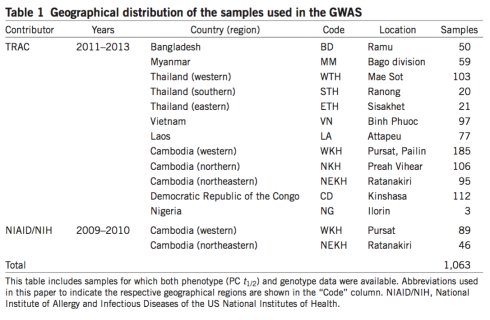
三、测序
Illumina测序平台。
paired-end sequencing reads of 200–300bp
1Gb of read data per sample
四:研究结果
1)Manhattan plot 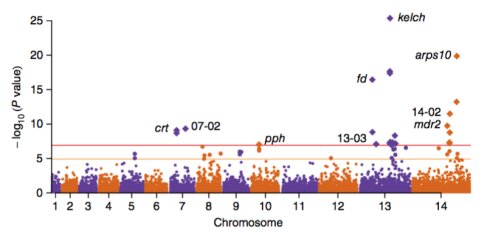
没有意义的SNP的都是圆点,有意义的都是菱形的,处于中间有意义的点,要比没有意义的点大。
其中这些SNP关联的基因如下: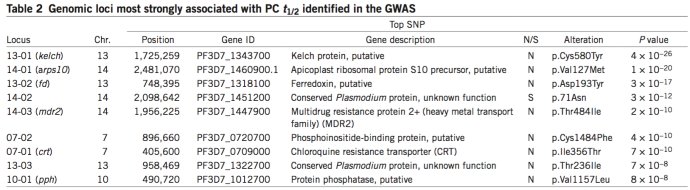
2)构建系统进化树,种群结构主要分为三个部分: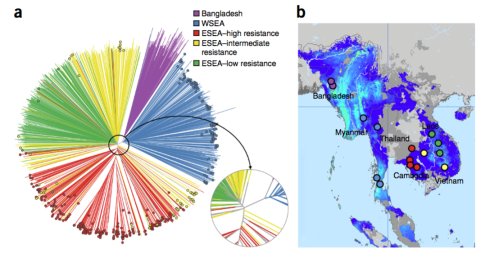
3)选择清楚分析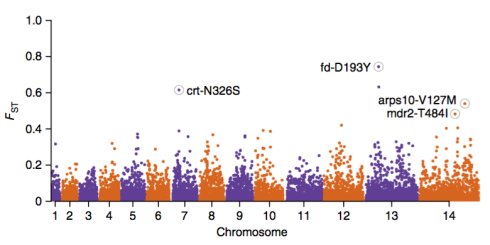
五数据分析
1)使用bwa进行比对
2)call snp使用的是samtools,总共得到3,373,632 SNP
3)生成的snp list使用SNP-o-matic algorithm 进行realignment 减少错误比对
(http://www.sanger.ac.uk/science/tools/snp-o-matic)
4)SNP过滤:
noncoding SNPs;
SNPs where polymorphisms had extremely low support (<10 reads in 1 sample);
SNPs with more than 2 alleles, with the exception of loci known to be important for drug resistance, which were manually verified to not have artifacts;
SNPs where coverage across samples was lower than the 25th percentile or higher than the 95th percentile of
(v) SNPs located in regions of relatively low uniqueness;
(vi) SNPs where heterozygosity levels were found to be inconsistent with the heterozygosity distribution at the SNP’s allele frequency;
(vii) SNPs where the genotype could not be established in at least 70% of samples.
5)关联分析使用:FaST-LMM v2.06 (因为样本多)
6)样本间关系矩阵计算使用snp的一个子集,主要是排除SNP当中的连锁的SNP,使用plink软件,参数为:–indep-pairwise 100 10 0.3 –maf 0.01
a) consider a window of 100 SNPs, b) calculate LD between each pair of SNPs in the window, b) remove one of a pair of SNPs if the LD is greater than 0.3, c) shift the window 10 SNPs forward and repeat the procedure.
(http://pngu.mgh.harvard.edu/~purcell/plink/summary.shtml#prune)
原文:http://blog.sina.com.cn/s/blog_83f77c940102w2wg.html
【GWAS文献解读】疟原虫青蒿素抗药性的全基因组关联分析
英文名:Genetic architecture of artemisinin-resistant Plasmodium falciparum
中文名:疟原虫青蒿素抗药性的全基因组关联分析