这本笔记本的目的是让你可视化各种分类器的决策边界。
本笔记本中使用的数据基于存储在mushrooms.csv中的UCI蘑菇数据集。
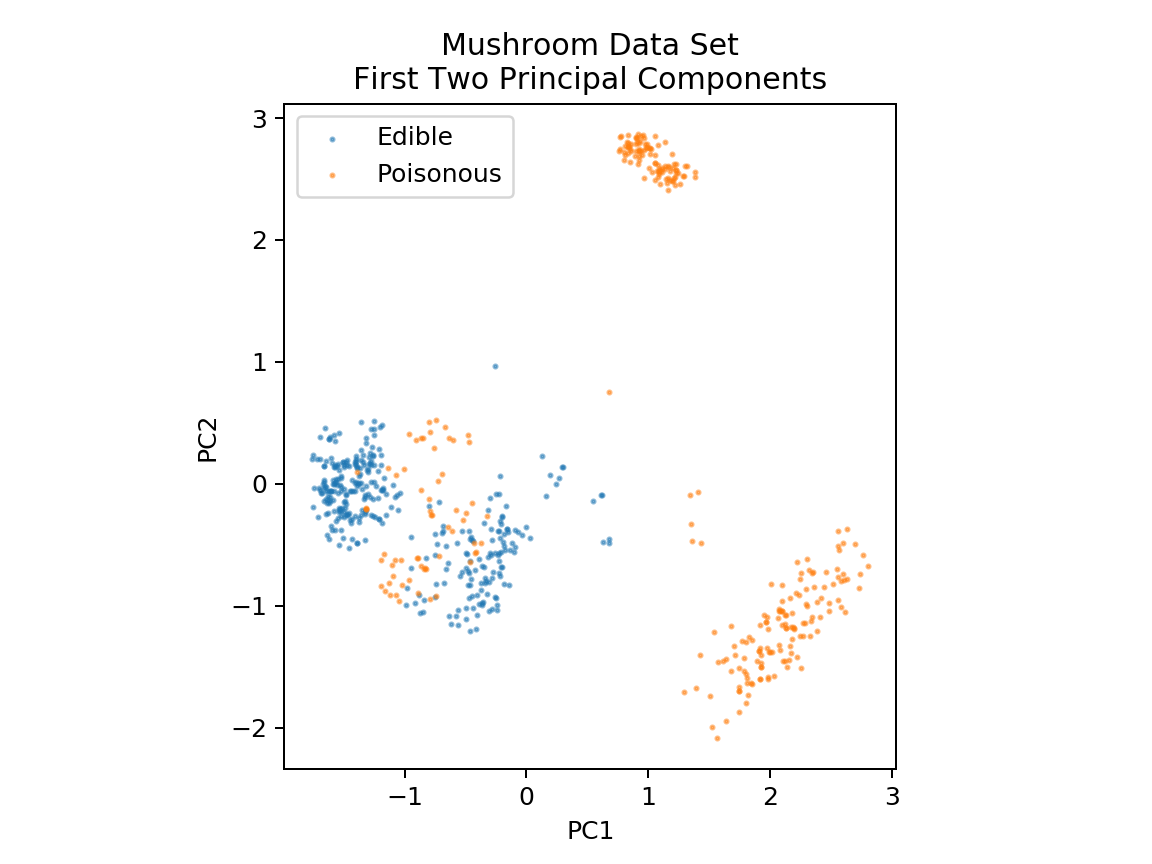
为了更好地确定决策边界,我们将对数据执行主成分分析(PCA)以将维度降至2维。 降维将在本课程后面的模块中介绍。
玩弄不同的模型和参数,看看它们如何影响分类器的决策边界和准确性!
1 %matplotlib notebook 2 3 import pandas as pd 4 import numpy as np 5 import matplotlib.pyplot as plt 6 from sklearn.decomposition import PCA 7 from sklearn.model_selection import train_test_split 8 9 df = pd.read_csv('mushrooms.csv') 10 df2 = pd.get_dummies(df) 11 12 df3 = df2.sample(frac=0.08) 13 14 X = df3.iloc[:,2:] 15 y = df3.iloc[:,1] 16 17 18 pca = PCA(n_components=2).fit_transform(X) 19 20 X_train, X_test, y_train, y_test = train_test_split(pca, y, random_state=0) 21 22 23 plt.figure(dpi=120) 24 plt.scatter(pca[y.values==0,0], pca[y.values==0,1], alpha=0.5, label='Edible', s=2) 25 plt.scatter(pca[y.values==1,0], pca[y.values==1,1], alpha=0.5, label='Poisonous', s=2) 26 plt.legend() 27 plt.title('Mushroom Data Set First Two Principal Components') 28 plt.xlabel('PC1') 29 plt.ylabel('PC2') 30 plt.gca().set_aspect('equal')
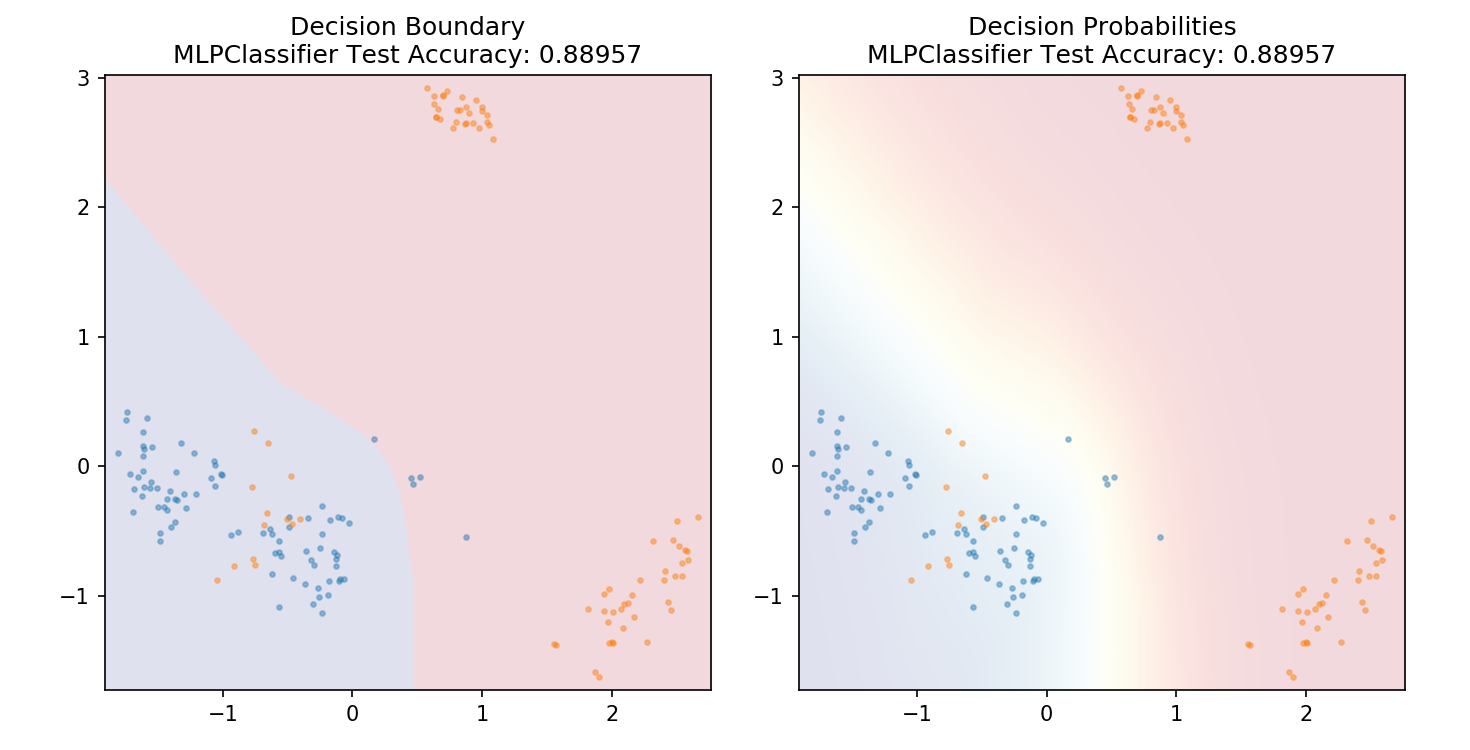
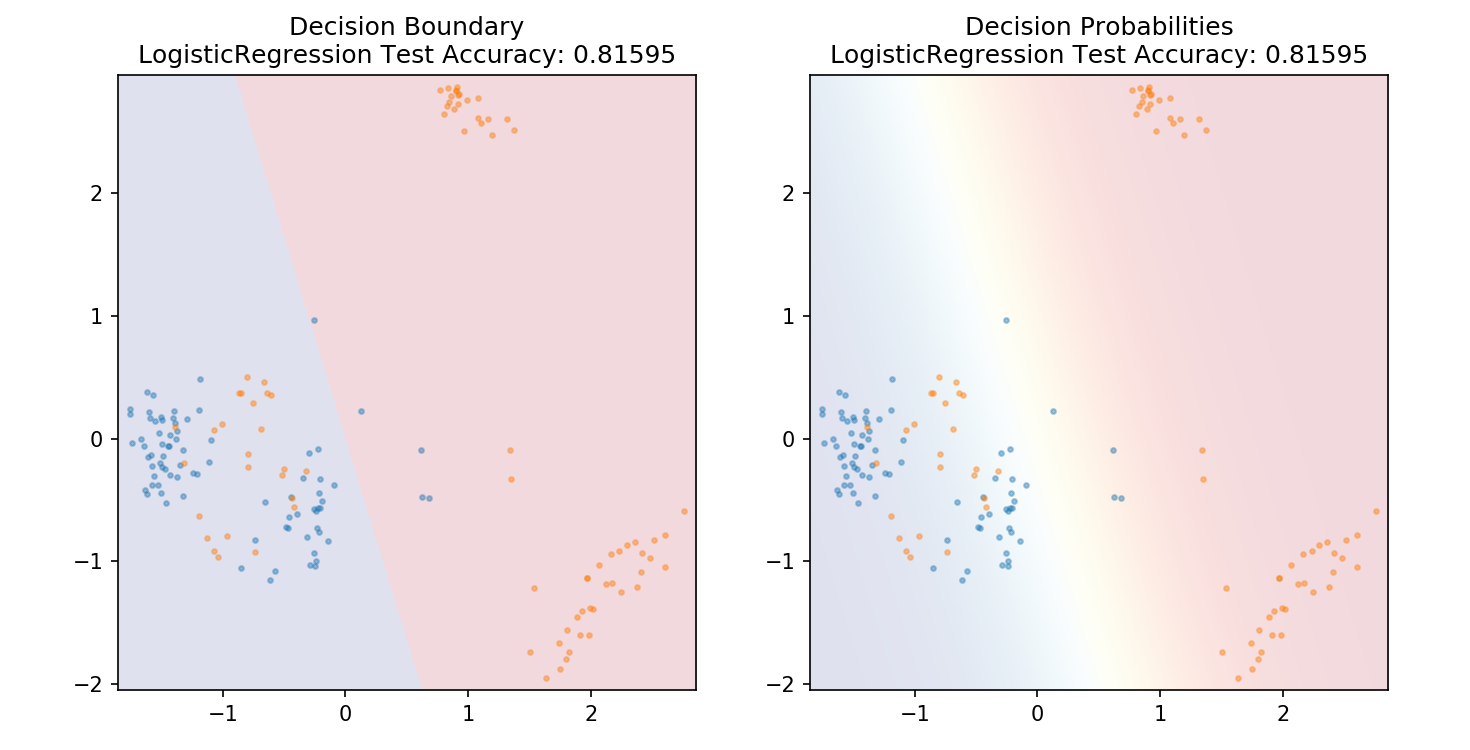
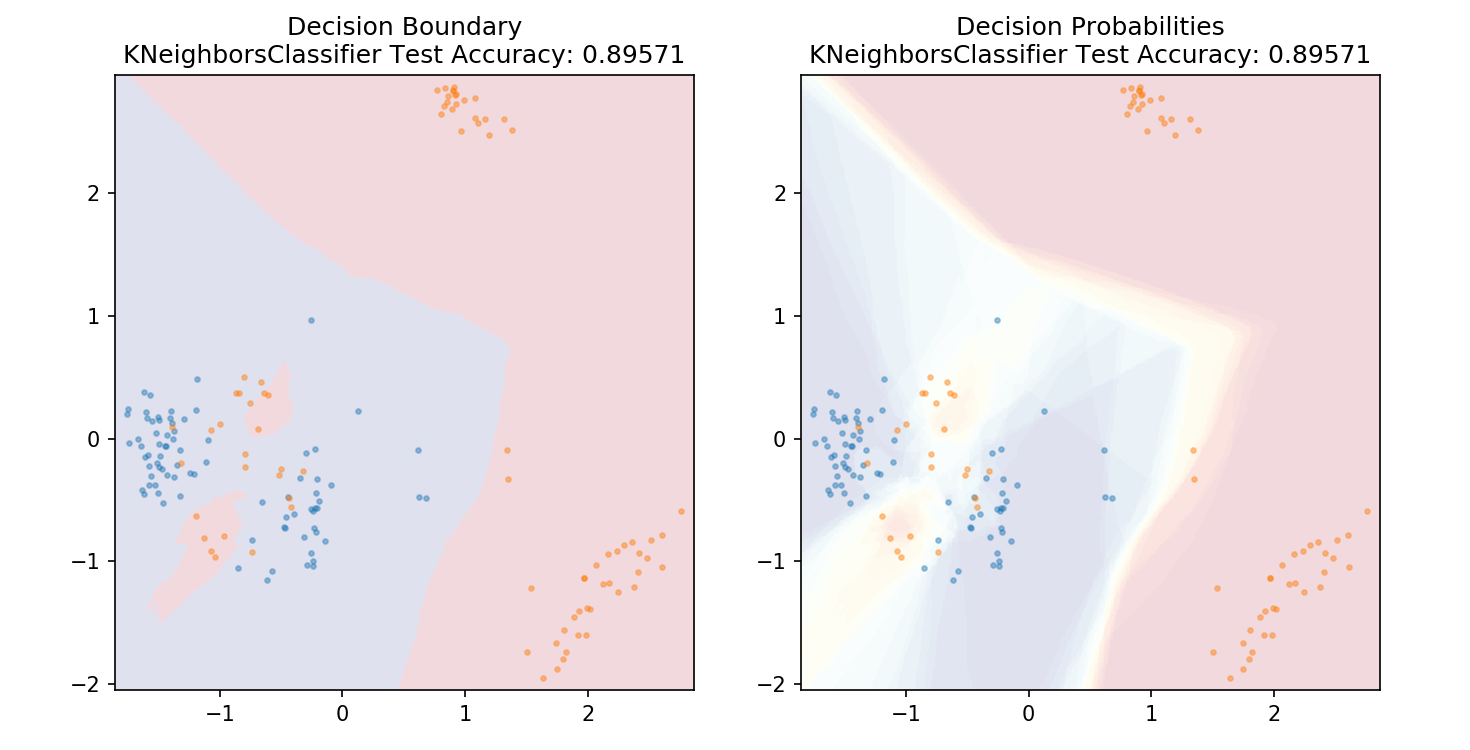
1 def plot_mushroom_boundary(X, y, fitted_model): 2 3 plt.figure(figsize=(9.8,5), dpi=100) 4 5 for i, plot_type in enumerate(['Decision Boundary', 'Decision Probabilities']): 6 plt.subplot(1,2,i+1) 7 8 mesh_step_size = 0.01 # step size in the mesh 9 x_min, x_max = X[:, 0].min() - .1, X[:, 0].max() + .1 10 y_min, y_max = X[:, 1].min() - .1, X[:, 1].max() + .1 11 xx, yy = np.meshgrid(np.arange(x_min, x_max, mesh_step_size), np.arange(y_min, y_max, mesh_step_size)) 12 if i == 0: 13 Z = fitted_model.predict(np.c_[xx.ravel(), yy.ravel()]) 14 else: 15 try: 16 Z = fitted_model.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:,1] 17 except: 18 plt.text(0.4, 0.5, 'Probabilities Unavailable', horizontalalignment='center', 19 verticalalignment='center', transform = plt.gca().transAxes, fontsize=12) 20 plt.axis('off') 21 break 22 Z = Z.reshape(xx.shape) 23 plt.scatter(X[y.values==0,0], X[y.values==0,1], alpha=0.4, label='Edible', s=5) 24 plt.scatter(X[y.values==1,0], X[y.values==1,1], alpha=0.4, label='Posionous', s=5) 25 plt.imshow(Z, interpolation='nearest', cmap='RdYlBu_r', alpha=0.15, 26 extent=(x_min, x_max, y_min, y_max), origin='lower') 27 plt.title(plot_type + ' ' + 28 str(fitted_model).split('(')[0]+ ' Test Accuracy: ' + str(np.round(fitted_model.score(X, y), 5))) 29 plt.gca().set_aspect('equal'); 30 31 plt.tight_layout() 32 plt.subplots_adjust(top=0.9, bottom=0.08, wspace=0.02)
1 from sklearn.linear_model import LogisticRegression 2 3 model = LogisticRegression() 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)
1 from sklearn.neighbors import KNeighborsClassifier 2 3 model = KNeighborsClassifier(n_neighbors=20) 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)
1 from sklearn.tree import DecisionTreeClassifier 2 3 model = DecisionTreeClassifier(max_depth=3) 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)
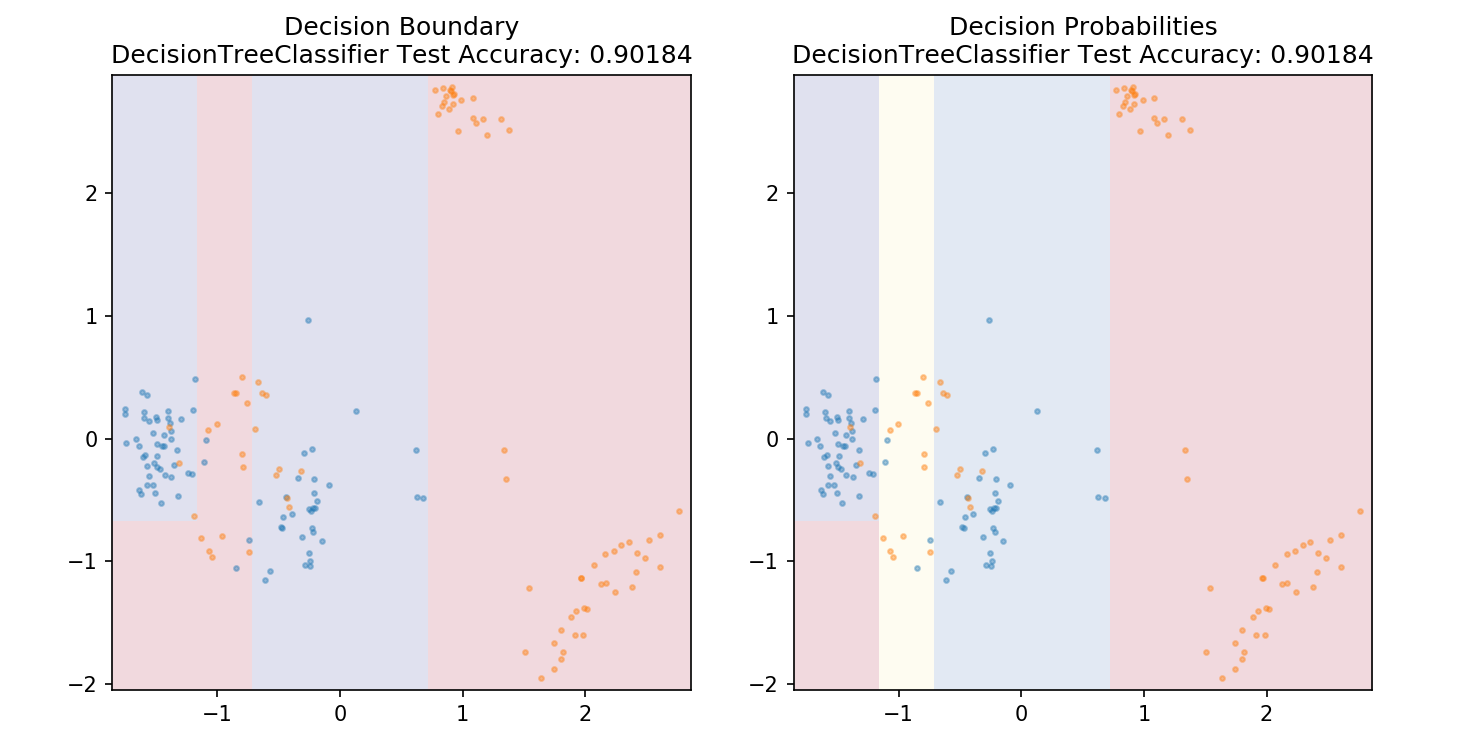
1 from sklearn.tree import DecisionTreeClassifier 2 3 model = DecisionTreeClassifier() 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)

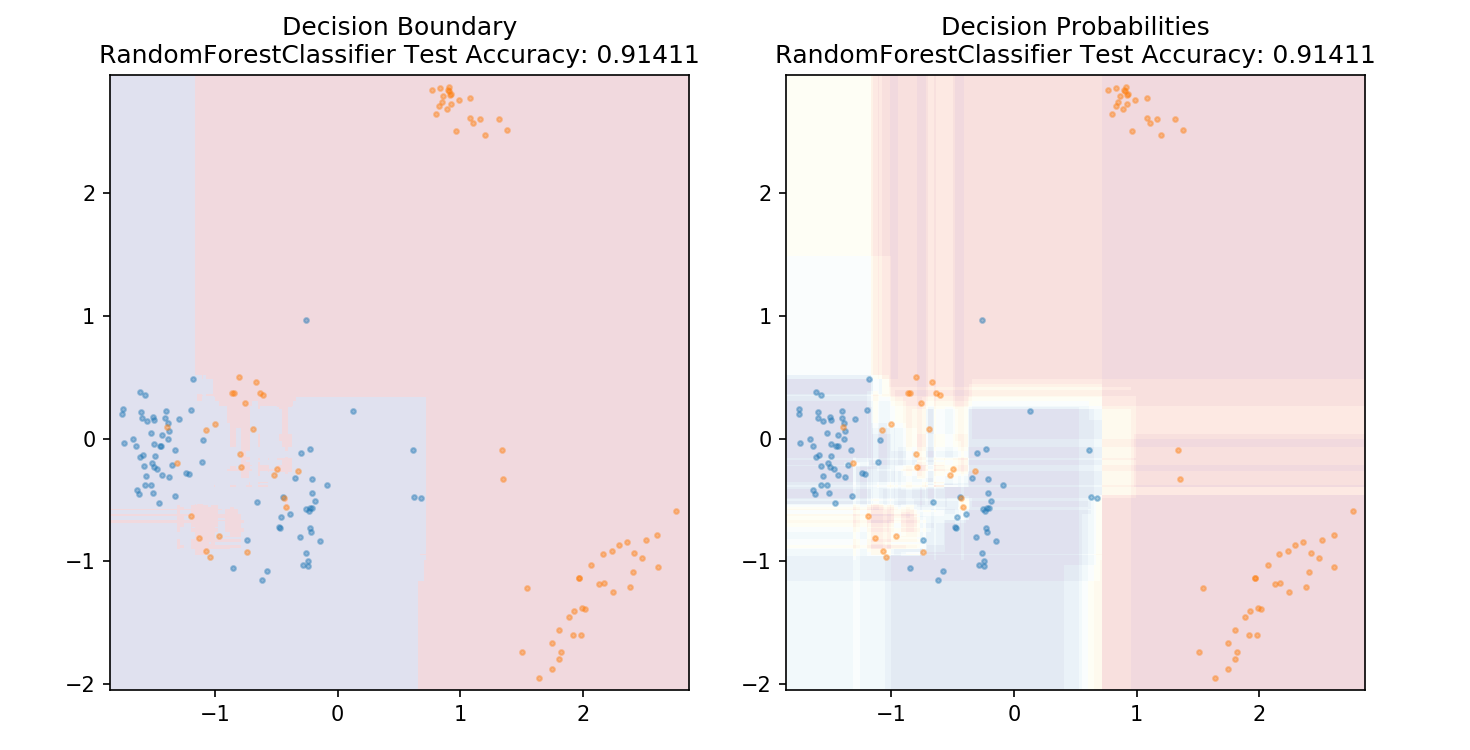
1 from sklearn.ensemble import RandomForestClassifier 2 3 model = RandomForestClassifier() 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)
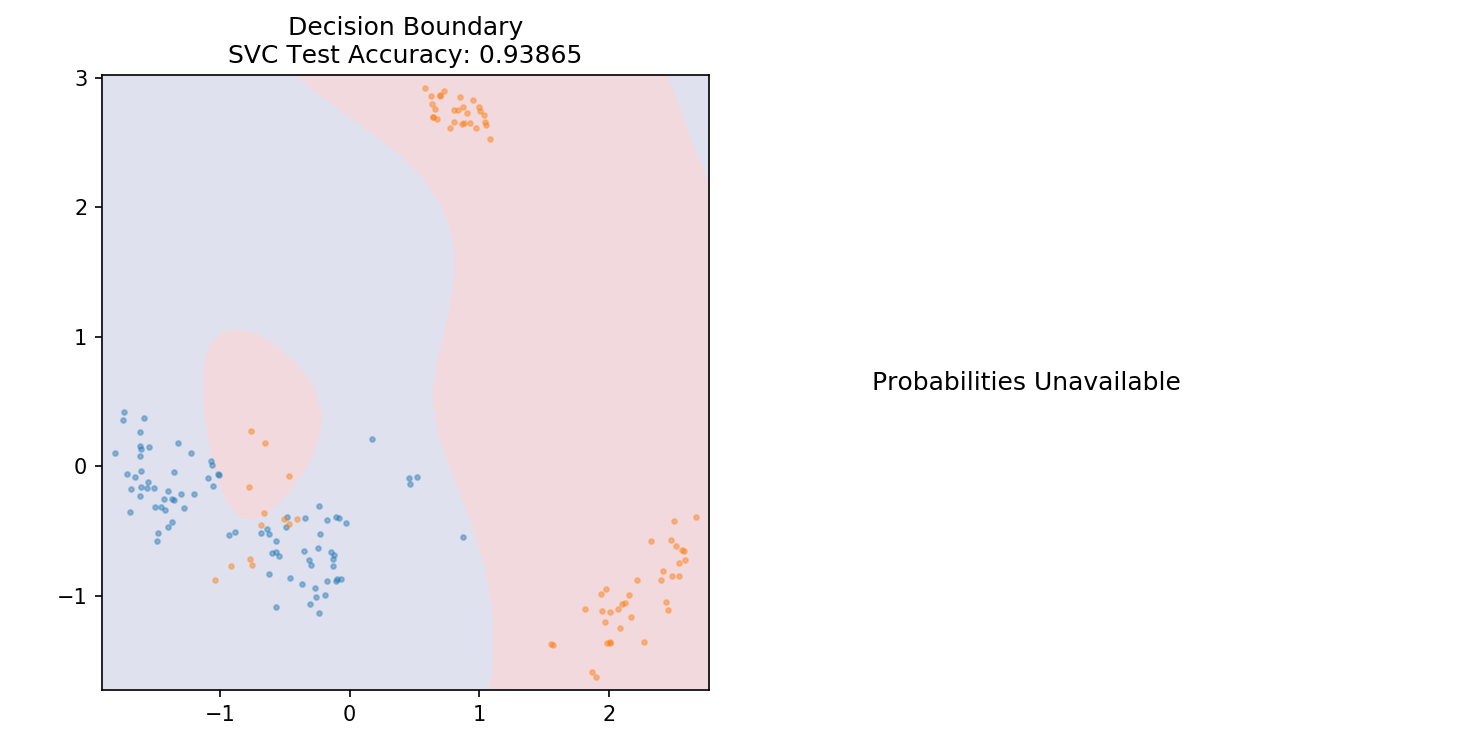
1 from sklearn.svm import SVC 2 3 model = SVC(kernel='linear') 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)

1 from sklearn.svm import SVC 2 3 model = SVC(kernel='rbf', C=1) 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)

1 from sklearn.svm import SVC 2 3 model = SVC(kernel='rbf', C=10) 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)
1 from sklearn.naive_bayes import GaussianNB 2 3 model = GaussianNB() 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)

1 from sklearn.neural_network import MLPClassifier 2 3 model = MLPClassifier() 4 model.fit(X_train,y_train) 5 6 plot_mushroom_boundary(X_test, y_test, model)