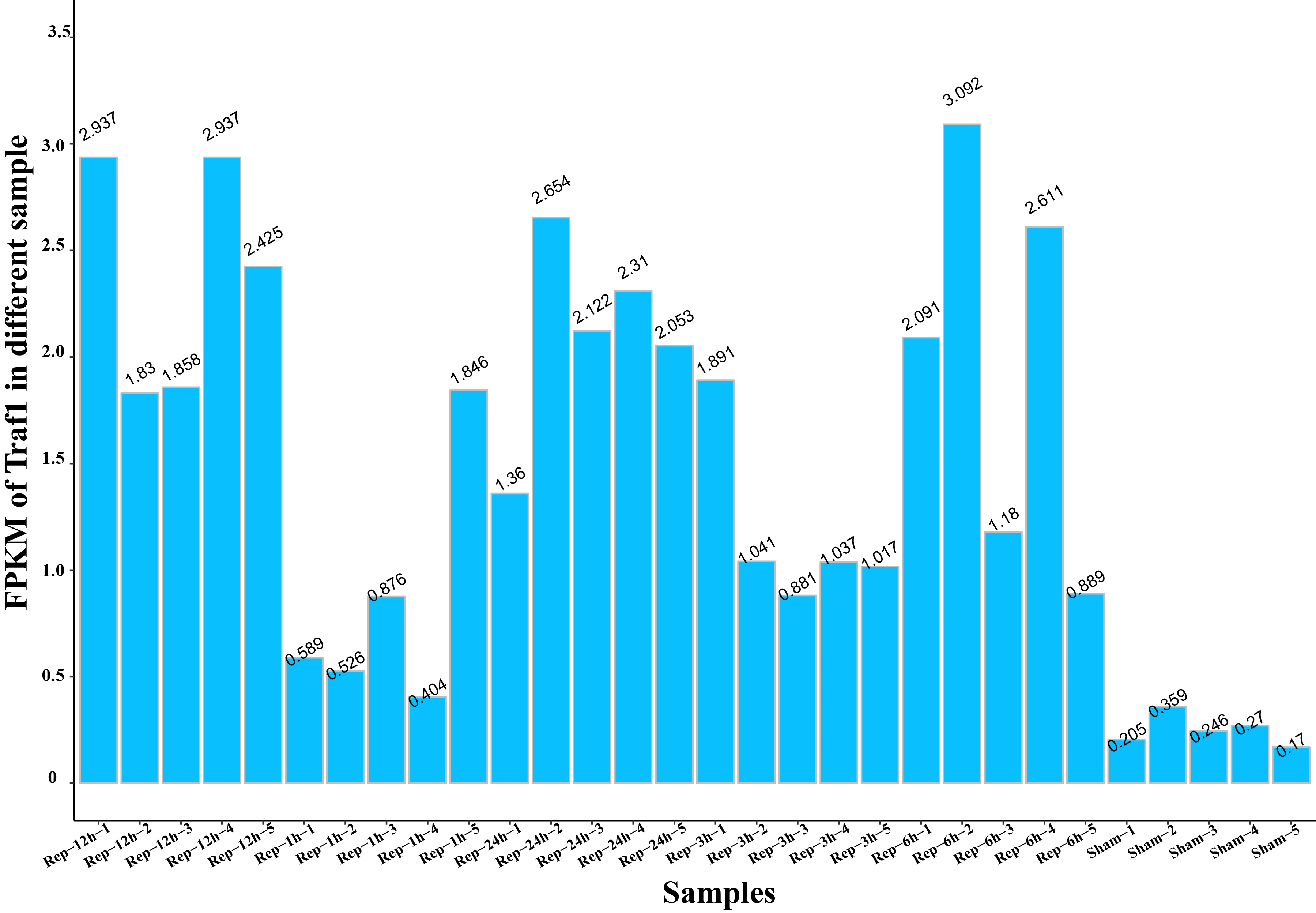
- 数据格式如下
gene_id Sham-1 Sham-2 Sham-3 Sham-4 Sham-5 Rep-1h-1 Rep-1h-2 Rep-1h-3 Rep-1h-4 Rep-1h-5 Rep-3h-1 Rep-3h-2 Rep-3h-3 Rep-3h-4 Rep-3h-5 Rep-6h-1 Rep-6h-2 Rep-6h-3 Rep-6h-4 Rep-6h-5 Rep-12h-1 Rep-12h-2 Rep-12h-3 Rep-12h-4 Rep-12h-5 Rep-24h-1 Rep-24h-2 Rep-24h-3 Rep-24h-4 Rep-24h-5
Traf1 0.204531 0.358811 0.24649 0.270231 0.169885 0.588808 0.526418 0.87557 0.403861 1.846186 1.890555 1.041459 0.881003 1.036722 1.016675 2.09069 3.09231 1.180259 2.610673 0.888904 2.936677 1.829962 1.857749 2.93743 2.424644 1.3602 2.654057 2.121849 2.309851 2.052516
- 代码如下:
a=read.table(file="Traf1.fpkm.lst")
c=t.data.frame(a)
gene_id=c(c[,1])
gene=as.vector(gene_id)
id=gene[2:length(gene)]
va=as.vector(c[,2])[2:length(as.vector(c[,2]))]
fpkm=round(as.numeric(va),3)
dd=data.frame(n,fpkm)
ggplot(dd,aes(x=n,y=fpkm))+
geom_bar(stat = "identity",fill ="#09BFFE",colour="grey")+
xlab("Samples")+ylab("FPKM of Traf1 in different sample")+
theme_classic()+
theme(axis.text.y=element_blank(),axis.text.x=element_text(size=8,color="black",angle = 30,face="bold",hjust = 1))+
geom_text(aes(label=dd$fpkm), position=position_stack(vjust=1.05),angle=30)+
scale_y_continuous(breaks = seq(0,3.5,0.5),limits = c(0,3.5),labels = seq(0,3.5,0.5))
使用fimo对Traf1基因进行motif的寻找
- 下载motif数据库
wget http://meme-suite.org/meme-software/Databases/motifs/motif_databases.12.18.tgz
tar zxvf motif_databases.12.18.tgz
代码如下
ln -s /media/sda/user/chengxu/project/cardiac-IR/lncRNA_sequencing/expression_data/mRNA_symbol_FPKM_matrix.txt ./
les mRNA_symbol_FPKM_matrix.txt|perl -F" " -lane 'print join(" ",@F[0..5])." ".join(" ",@F[43..67])' |head -1 >Traf1.fpkm.lst
les mRNA_symbol_FPKM_matrix.txt|perl -F" " -lane 'print join(" ",@F[0..5])." ".join(" ",@F[43..67])' |grep Traf1 >>Traf1.fpkm.lst
grep Traf1 /media/sda/database/Ensembl/release-89/mus_musculus/gtf/gene.bed.txt |perl -lane '$s=$F[5]-1;$e=$F[6]-1;print qq{$F[3] $s $e $F[7]}' >Traf1.bed
bedtools getfasta -fi /media/sda/database/Ensembl/release-89/mus_musculus/fasta/mm10.fasta -bed Traf1.bed -fo Traf1.gene.fasta
fimo -oc Traf1 JASPAR2018_CORE_vertebrates_non-redundant.meme Traf1.gene.fasta
- 需要注意的是bed文件时从0开始算位置的,而gtf等注释文件时从1开始算的,所以gtf转换成bed的时候要减少一个碱基的位置
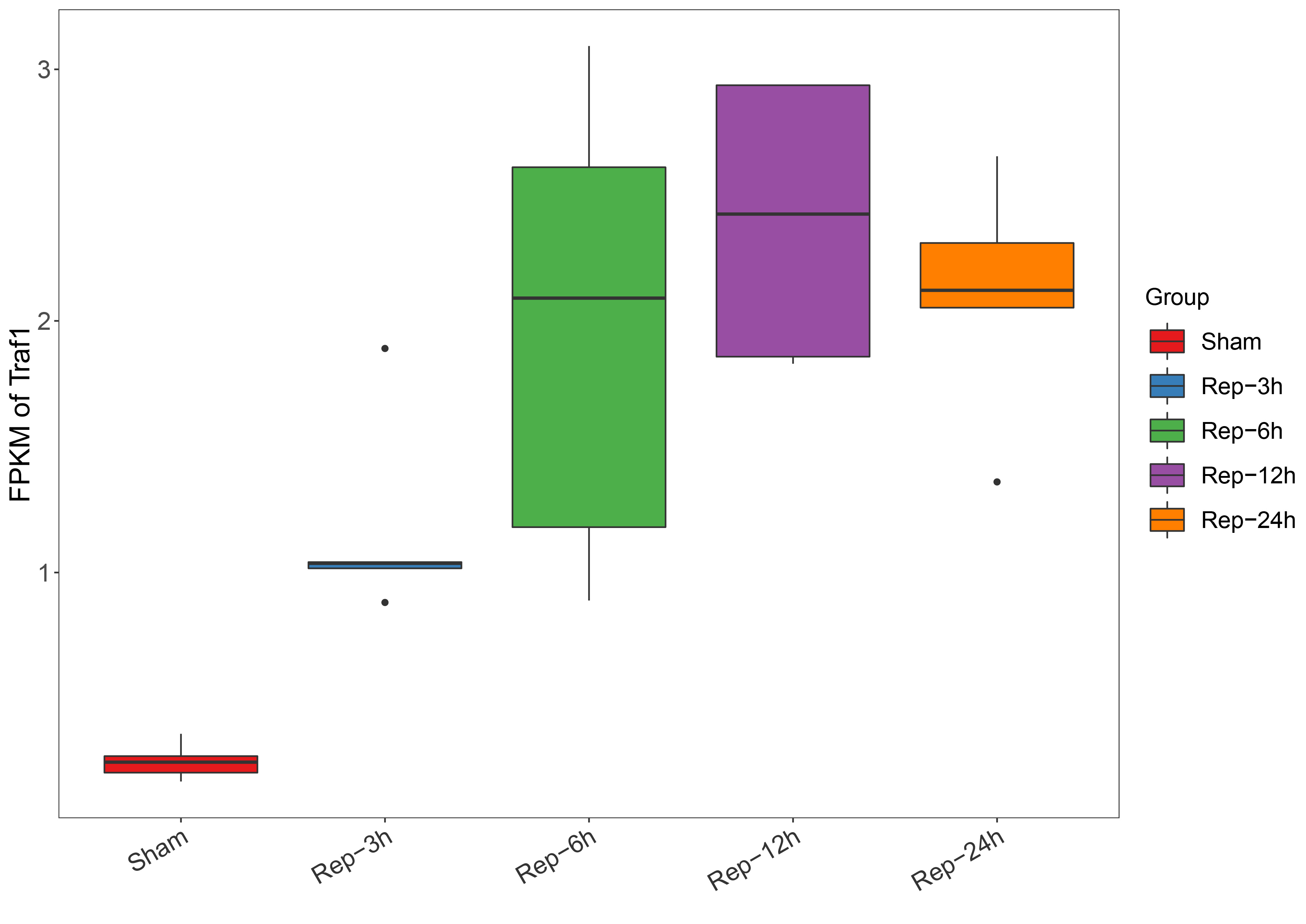
- 数据格式
aaa sss fpkm
Sham Sham-1 0.204531
Sham Sham-2 0.358811
Sham Sham-3 0.24649
Sham Sham-4 0.270231
Sham Sham-5 0.169885
Rep-1h Rep-1h-1 0.588808
Rep-1h Rep-1h-2 0.526418
Rep-1h Rep-1h-3 0.87557
Rep-1h Rep-1h-4 0.403861
Rep-1h Rep-1h-5 1.846186
Rep-3h Rep-3h-1 1.890555
Rep-3h Rep-3h-2 1.041459
Rep-3h Rep-3h-3 0.881003
Rep-3h Rep-3h-4 1.036722
Rep-3h Rep-3h-5 1.016675
Rep-6h Rep-6h-1 2.09069
Rep-6h Rep-6h-2 3.09231
Rep-6h Rep-6h-3 1.180259
Rep-6h Rep-6h-4 2.610673
Rep-6h Rep-6h-5 0.888904
Rep-12h Rep-12h-1 2.936677
Rep-12h Rep-12h-2 1.829962
Rep-12h Rep-12h-3 1.857749
Rep-12h Rep-12h-4 2.93743
Rep-12h Rep-12h-5 2.424644
Rep-24h Rep-24h-1 1.3602
Rep-24h Rep-24h-2 2.654057
Rep-24h Rep-24h-3 2.121849
Rep-24h Rep-24h-4 2.309851
Rep-24h Rep-24h-5 2.052516
- 画图代码
dd=read.table(file = "Traf1.changeformat",header = T)
#修改箱线图的顺序
dd$aaa=factor(dd$aaa,levels=c("Sham","Rep-3h","Rep-6h","Rep-12h","Rep-24h"))
ggplot(dd, aes(x=aaa, y=fpkm, fill=aaa)) +
geom_boxplot() +
scale_fill_manual(values=c("#E41A1C","#377EB8","#4DAF4A","#984EA3","#FF7F00","#FFFF33"), name="Var2") +
labs(x="", y="FPKM of Traf1") + #去掉x轴的Var2
guides(fill = guide_legend(title="Group", keywidth=2, keyheight=2)) +
theme_bw() +
theme(panel.grid = element_blank()) +
theme(axis.title.y = element_text(size=18)) +
theme(axis.text.y = element_text(size=16, hjust=1)) +
theme(axis.text.x = element_text(colour="grey20", size=96/len, angle=30, hjust=1)) +
theme(legend.title = element_text(size=15)) +
theme(legend.text = element_text(size=15))