1 基于随机森林的医疗费用分析与建模预估
import warnings warnings.filterwarnings('ignore') # 忽视警告
import pandas as pd from matplotlib import pyplot as plt import seaborn as sns from sklearn.ensemble import RandomForestRegressor from sklearn.preprocessing import LabelEncoder, StandardScaler from sklearn.model_selection import train_test_split import numpy as np import sklearn.metrics
#1.加载数据,并进行分析¶ df = pd.read_csv('./data/insurance.csv') #剔除缺失值 df = df.dropna() df.head()

df.info()
<class 'pandas.core.frame.DataFrame'> Int64Index: 1338 entries, 0 to 1337 Data columns (total 7 columns): age 1338 non-null int64 sex 1338 non-null object bmi 1338 non-null float64 children 1338 non-null int64 smoker 1338 non-null object region 1338 non-null object charges 1338 non-null float64 dtypes: float64(2), int64(2), object(3) memory usage: 83.6+ KB
#查看数据分布状况 df.describe() # 从年龄能看出,平均在39岁

age:年龄 sex:性别 bmi:身体质量指数 children:zi smoker region charges
#查看不同维度之间的相关性,倒置的手法 df.corr()

# 注:bmi(Body Mass Index)为身体质量指数,这里省略小数点来区间化特征,方便做数据分析 df['bmi_int'] = df['bmi'].apply(lambda x: int(x)) variables=['sex','smoker','region','age','bmi_int','children'] print('数据分布分析:') """ 参数 说明 by 指定列名(axis=0或’index’)或索引值(axis=1或’columns’) axis 若axis=0或’index’,则按照指定列中数据大小排序;若axis=1或’columns’,则按照指定索引中数据大小排序,默认axis=0 ascending 是否按指定列的数组升序排列,默认为True,即升序排列 inplace 是否用排序后的数据集替换原来的数据,默认为False,即不替换 na_position {‘first’,‘last’},设定缺失值的显示位置 """ for v in variables: df = df.sort_values(by=[v]) df[v].value_counts().plot(kind = 'bar') plt.title(v) plt.show()
数据分布分析:

- 0:代表没孩子 1:代表1个孩子......
#对类别型变量进行编码 le_sex = LabelEncoder() le_smoker = LabelEncoder() le_region = LabelEncoder() df['sex'] = le_sex.fit_transform(df['sex']) df['smoker'] = le_smoker.fit_transform(df['smoker']) df['region'] = le_region.fit_transform(df['region']) # 0 1 2 3
df['region']
172 0
1150 0
195 1
581 1
1196 1
..
937 1
425 2
1245 3
1085 3
932 3
Name: region, Length: 1338, dtype: int64
variables=['sex','smoker','region','age','bmi','children'] # 选择特征 X=df[variables] # 导入特征数据 sc=StandardScaler() X=sc.fit_transform(X) # 对数据进行标准化处理 Y=df['charges'] # 导入标签数据 # 使用sklearn.model_selection库自动对数据进行训练集与测试集的分割 X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
# 使用import的随机森林模型,并设置不同的树的数量进行试验,参考值为200 regressor = RandomForestRegressor(n_estimators=200) # 使用fit函数对X_train,y_train进行拟合 regressor.fit(X_train,y_train) #prediction and evaluation y_train_pred = regressor.predict(X_train) #对X_train进行预测 y_test_pred = regressor.predict(X_test) # 对X_test进行预测
#计算相应的MAE和RMSE,得到价格的平均绝对值损失 print('RandomForestRegressor evaluating result:') print("Train MAE: ", sklearn.metrics.mean_absolute_error(y_train ,y_train_pred )) print("Train RMSE: ", np.sqrt(sklearn.metrics.mean_squared_error(y_train,y_train_pred))) print("Test MAE: ", sklearn.metrics.mean_absolute_error(y_test ,y_test_pred)) print("Test RMSE: ", np.sqrt(sklearn.metrics.mean_squared_error(y_test, y_test_pred)))
RandomForestRegressor evaluating result: Train MAE: 944.9813798049196 Train RMSE: 1764.1825891298656 Test MAE: 2993.187358598303 Test RMSE: 5603.299343502578
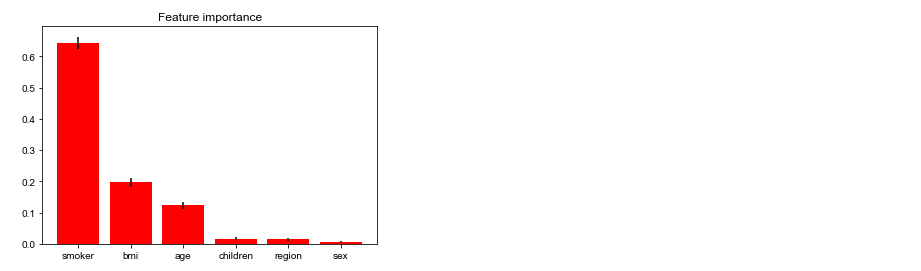
# 简单可视化模型,查看各个特征在随机森林中的重要度 importances = regressor.feature_importances_ std = np.std([tree.feature_importances_ for tree in regressor.estimators_],axis=0)# 代表行 indices = np.argsort(importances)[::-1] importance_list = [] # shape[1]:代表一维矩阵 for f in range(X.shape[1]): variable = variables[indices[f]] # index,就是序列号 importance_list.append(variable) print("%d.%s(%f)" % (f + 1, variable, importances[indices[f]])) # Plot the feature importances of the forest plt.figure() plt.title("Feature importance") plt.bar(importance_list, importances[indices], color="r", yerr=std[indices], align="center") plt.show()
特征重要度排序 1.smoker(0.643205) 2.bmi(0.197045) 3.age(0.123219) 4.children(0.016878) 5.region(0.014015) 6.sex(0.005639)