1. 计算每一种的比例的百分比
import pandas as pd from matplotlib.ticker import FuncFormatter np.random.seed(0) df = pd.DataFrame({'Condition 1':np.random.rand(20), 'Condition 2':np.random.rand(20)*0.9, 'Condtion 3':np.random.rand(20)*1.1}) print(df.head()) fig, ax = plt.subplots() # stacked 进行堆叠操作 df.plot.bar(ax=ax, stacked=True) plt.show() # 设置百分比 df_ratio = df.div(df.sum(axis=1), axis=0) fig, ax = plt.subplots() df_ratio.plot.bar(ax=ax, stacked=True) ax.yaxis.set_major_formatter(FuncFormatter(lambda y,_:'{:.0%}'.format(y))) plt.show()


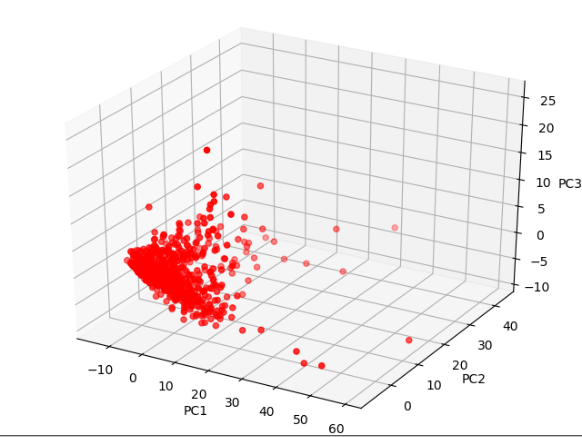
2. 通过pd将数据导入,进行缺失值补充,画出特征的PCA图
# 1 下载数据 url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/00383/risk_factors_cervical_cancer.csv' df = pd.read_csv(url, na_values='?') print(df.head()) # 2.对缺失值进行补充 from sklearn.preprocessing import Imputer # 引入inputer() 使用均值对缺失值进行填充 impute = pd.DataFrame(Imputer().fit_transform(df)) print(impute.head()) impute.columns = df.columns impute.index = df.index import seaborn as sns from sklearn.decomposition import PCA from mpl_toolkits.mplot3d import Axes3D # 3.取出样品特征, 取出Dx:Cancer features = impute.drop('Dx:Cancer', axis=1) y = impute['Dx:Cancer'] # 4进行PCA操作 pca = PCA(n_components=3) X_r = pca.fit_transform(features) # '{:.2%}'表示保留两位小数, pca.explained_variabce_ratio表示所占的比例 print('Explained variance: PC1{:.2%} PC2{:.2%} PC3{:.2%}' .format(pca.explained_variance_ratio_[0], pca.explained_variance_ratio_[1], pca.explained_variance_ratio_[2],)) # 构造三维坐标系 fig = plt.figure() ax = Axes3D(fig) # 画散点图 ax.scatter(X_r[:, 0], X_r[:, 1], X_r[:, 2], c='r', cmap=plt.cm.coolwarm) # 对三个维度的坐标进行标注 ax.set_xlabel('PC1') ax.set_ylabel('PC2') ax.set_zlabel('PC3') plt.show()