1、测试数据
root@PC1:/home/test# ls 5gene_CDs.bed id_mapping.txt root@PC1:/home/test# head -n 3 5gene_CDs.bed chr6 117609654 117609965 NM_001378891.1_cds_0_0_chr6_117609655_r 0 - chr6 117622136 117622300 NM_001378891.1_cds_1_0_chr6_117622137_r 0 - chr6 117629956 117630091 NM_001378891.1_cds_2_0_chr6_117629957_r 0 - root@PC1:/home/test# head -n 3 id_mapping.txt ## 在5gene_CDs.bed文件中实现id_mapping.txt中第二列对第一列的批量替换 NM_001378891.1 ROS1 NM_001378902.1 ROS1 NM_002944.3 ROS1
2、shell实现
root@PC1:/home/test# ls 5gene_CDs.bed id_mapping.txt root@PC1:/home/test# head -n 3 5gene_CDs.bed chr6 117609654 117609965 NM_001378891.1_cds_0_0_chr6_117609655_r 0 - chr6 117622136 117622300 NM_001378891.1_cds_1_0_chr6_117622137_r 0 - chr6 117629956 117630091 NM_001378891.1_cds_2_0_chr6_117629957_r 0 - root@PC1:/home/test# head -n 3 id_mapping.txt NM_001378891.1 ROS1 NM_001378902.1 ROS1 NM_002944.3 ROS1 root@PC1:/home/test# cp 5gene_CDs.bed 5gene_CDs.bed.bak ## 要在源文件中修改,防止以外发生对数据做备份 root@PC1:/home/test# ls 5gene_CDs.bed 5gene_CDs.bed.bak id_mapping.txt root@PC1:/home/test# cat id_mapping.txt | while read {i,j}; do sed -i "s/$i/$j/" 5gene_CDs.bed; done ## 循环中i和j分别存储每行中对应的两个变量 root@PC1:/home/test# head -n 3 5gene_CDs.bed chr6 117609654 117609965 ROS1_cds_0_0_chr6_117609655_r 0 - chr6 117622136 117622300 ROS1_cds_1_0_chr6_117622137_r 0 - chr6 117629956 117630091 ROS1_cds_2_0_chr6_117629957_r 0 -
3、R实现
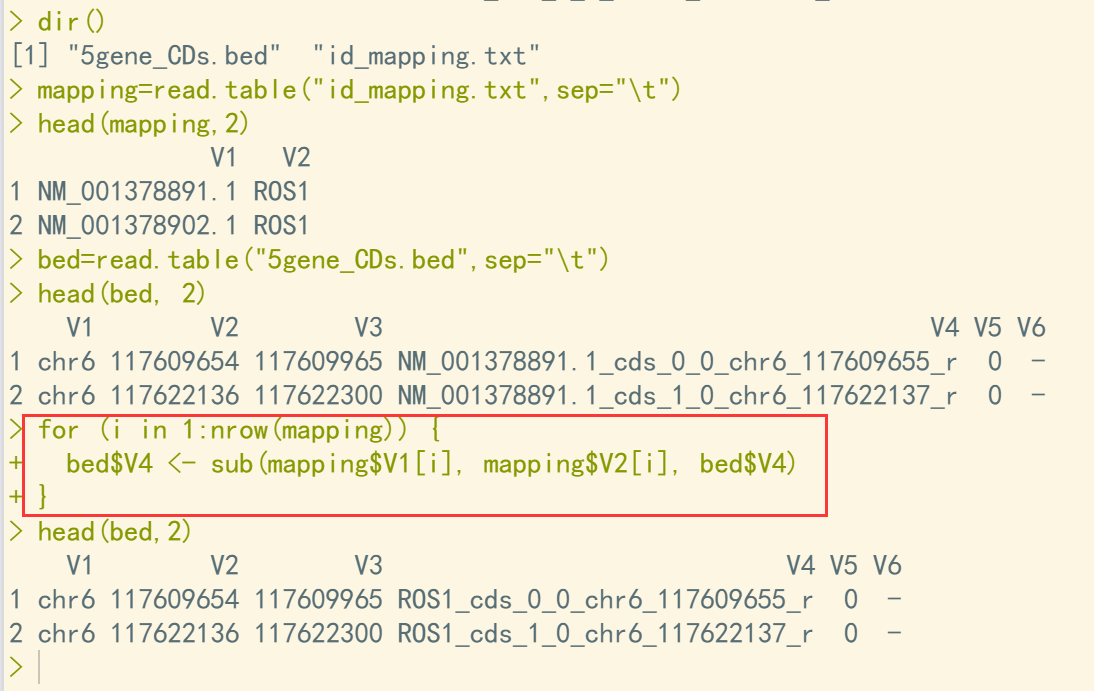
> dir() [1] "5gene_CDs.bed" "id_mapping.txt" > mapping=read.table("id_mapping.txt",sep="\t") > head(mapping,2) V1 V2 1 NM_001378891.1 ROS1 2 NM_001378902.1 ROS1 > bed=read.table("5gene_CDs.bed",sep="\t") > head(bed, 2) V1 V2 V3 V4 V5 V6 1 chr6 117609654 117609965 NM_001378891.1_cds_0_0_chr6_117609655_r 0 - 2 chr6 117622136 117622300 NM_001378891.1_cds_1_0_chr6_117622137_r 0 - > for (i in 1:nrow(mapping)) { + bed$V4 <- sub(mapping$V1[i], mapping$V2[i], bed$V4) + } > head(bed,2) V1 V2 V3 V4 V5 V6 1 chr6 117609654 117609965 ROS1_cds_0_0_chr6_117609655_r 0 - 2 chr6 117622136 117622300 ROS1_cds_1_0_chr6_117622137_r 0 -
测试数据来源: https://mp.weixin.qq.com/s?__biz=MzI4ODE0NTE3OA==&mid=2649219537&idx=1&sn=6974c6e3b91b6e91ac407563f10d3835&chksm=f3d1aba4c4a622b243e58434d561e2a08db148fb98499eaec626f6e186ec5cfa912c6db5ab93&mpshare=1&scene=23&srcid=1118L4C0wtH1cYHmjtZD2lrk&sharer_sharetime=1637166249484&sharer_shareid=4ed060cc4cd1efce40e3ab6dd8d8c7d4#rd