1、脚本
[root@centos79 test]# cat test.sh #!/bin/bash #step1 check ped file uniqn=$(sed 's/\r//g' $1 | cut -d " " -f 7- | sed 's/ /\n/g' | sort -u | wc -l) if [ $uniqn -gt 5 ] then echo "error, exception nucleotide: " sed 's/^M//g' $1 | cut -d " " -f 7- | sed 's/ /\n/g' | sort -u | grep -v [ATGC0] exit fi temp1=$(head -n 1 $1 | awk '{print NF}') for i in $(seq `sed -n "$=" $1`) do temp2=$(sed -n "$i"p $1 | awk '{print NF}') if [ $temp2 -ne $temp1 ] then echo "inconsistent number of columns!" echo -e "colun1 1: $temp1\ncolume $i: $temp2" exit fi done echo "step 1 done!" #step2 check consistence of ped and map file mapline=$(sed -n "$=" $2) locinum=$(head -n 1 $1 | awk '{print (NF - 6)/2}') if [ $mapline -ne $locinum ] then echo "error, map file and ped file do not match!" echo -e "mapline: $mapline\nlocinum: $locinum" exit fi echo "step 2 done!" #step 3 statistics lines=$(sed -n "$=" $1) for i in $(seq `head -n 1 $1 | awk '{print (NF - 6)/2}'`); do awk -v a=$i '{print $(a * 2 + 5), $(a * 2 + 6)}' $1 | awk -v a=$lines -v RS="@#$j" '{print gsub(/0/,"&")/(a*2)}' >> missresult1; done echo "step 3 done!" #step 4 add loci and headline cut -f 1-2,4 $2 | paste - missresult1 | sed '1i chr\tsnp\tpos\tmissing_rate' > missresult.txt rm -f missresult1 echo "fineshed!"
2、测试数据及用法
[root@centos79 test]# ls outcome.map outcome.ped test test.sh [root@centos79 test]# cat outcome.map 1 snp1 0 55910 1 snp2 0 85204 1 snp3 0 122948 1 snp4 0 203750 1 snp5 0 312707 1 snp6 0 356863 1 snp7 0 400518 1 snp8 0 487423 [root@centos79 test]# cat outcome.ped DOR 1 0 0 0 -9 G G 0 0 0 0 0 0 A A T T G G 0 0 DOR 2 0 0 0 -9 0 0 G G 0 0 0 0 A A T T A G 0 0 DOR 3 0 0 0 -9 0 0 C C 0 0 G G G G T T A G 0 0 DOR 4 0 0 0 -9 0 0 C C 0 0 G G G G A A G G G G DOR 5 0 0 0 -9 G G C C 0 0 G G G G A A A G G C DOR 6 0 0 0 -9 G G C C 0 0 G G G G A A A A C C [root@centos79 test]# bash test.sh outcome.ped outcome.map step 1 done! step 2 done! step 3 done! fineshed! [root@centos79 test]# ls missresult.txt outcome.map outcome.ped test test.sh [root@centos79 test]# cat missresult.txt chr snp pos missing_rate 1 snp1 55910 0.5 1 snp2 85204 0.166667 1 snp3 122948 1 1 snp4 203750 0.333333 1 snp5 312707 0 1 snp6 356863 0 1 snp7 400518 0 1 snp8 487423 0.5
3、plink软件验证
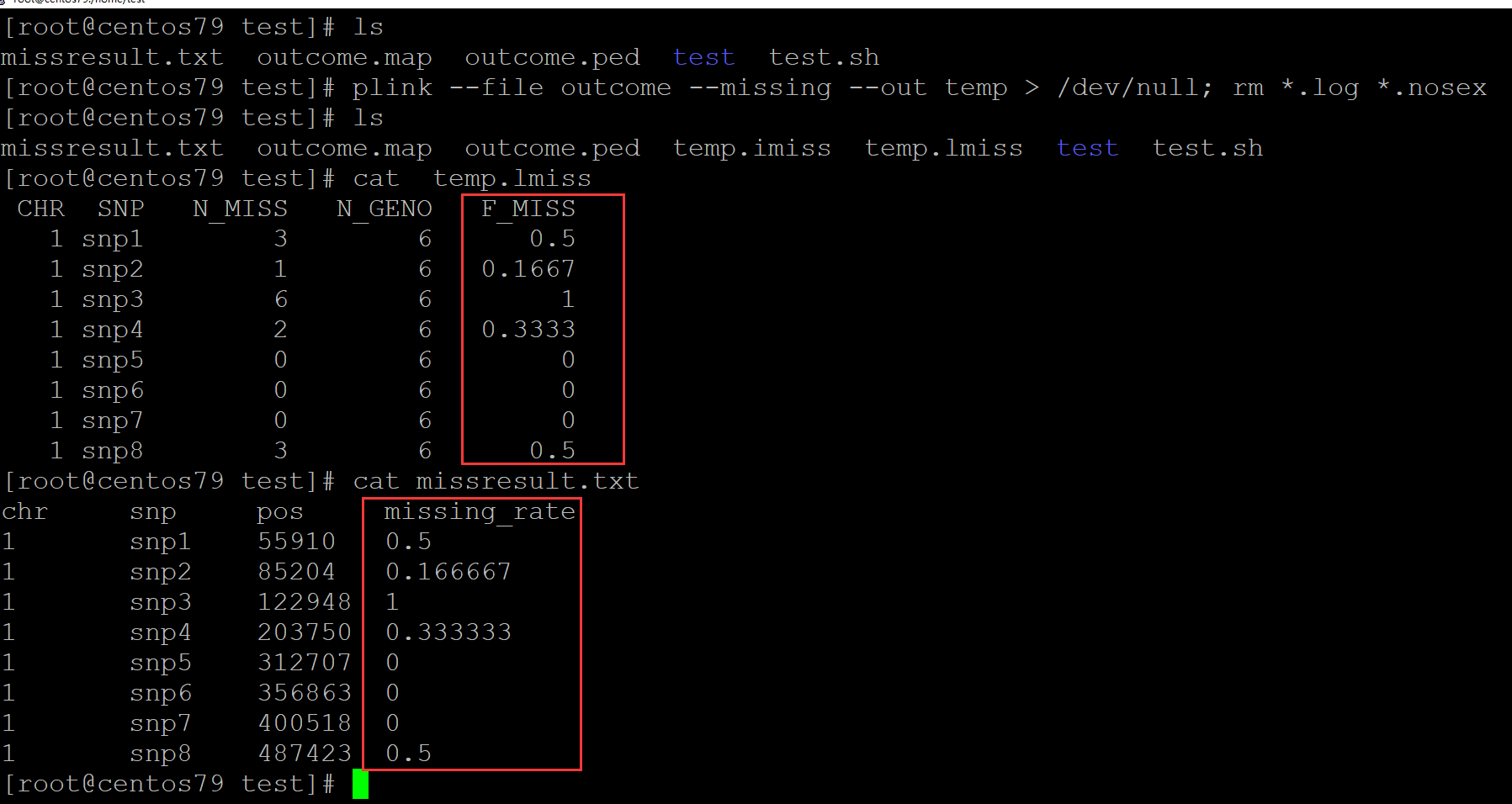
[root@centos79 test]# ls missresult.txt outcome.map outcome.ped test test.sh [root@centos79 test]# plink --file outcome --missing --out temp > /dev/null; rm *.log *.nosex [root@centos79 test]# ls missresult.txt outcome.map outcome.ped temp.imiss temp.lmiss test test.sh [root@centos79 test]# cat temp.lmiss CHR SNP N_MISS N_GENO F_MISS 1 snp1 3 6 0.5 1 snp2 1 6 0.1667 1 snp3 6 6 1 1 snp4 2 6 0.3333 1 snp5 0 6 0 1 snp6 0 6 0 1 snp7 0 6 0 1 snp8 3 6 0.5 [root@centos79 test]# cat missresult.txt chr snp pos missing_rate 1 snp1 55910 0.5 1 snp2 85204 0.166667 1 snp3 122948 1 1 snp4 203750 0.333333 1 snp5 312707 0 1 snp6 356863 0 1 snp7 400518 0 1 snp8 487423 0.5