1.参考 reference
1. tutorial主页:http://www.bcs.rochester.edu/people/raizada/fmri-matlab.htm。
2.speech_brain_images.mat数据:speech_brain_images.mat。
3.showing_brain_images_tutorial显示大脑图像代码:showing_brain_images_tutorial.m 。
4.overlaying_Tmaps_tutorial.m叠加t检验统计图:overlaying_Tmaps_tutorial.m。
2.程序执行过程 run step by step
2.1 显示大脑axial 轴位 第18个切片的图像,以灰度图的方式显示
clear all;
close all;
load speech_brain_images.mat
whos
%%%% This is the output that you should get:
%
% Name Size Bytes Class
%
% speech_Tmap 53x63x46 1228752 double array
% subj_3danat 53x63x46 1228752 double array
%%%% Let's look at the 18th axial slice.
%%%% This goes through Heschl's gyrus, which is auditory cortex
axial_slice_number = 18;
axial_slice_vals = subj_3danat(:,:,axial_slice_number);
%%%% Let's look at this matrix in a figure
figure(1);
clf;
imagesc(axial_slice_vals);
colormap gray; %%% Show the image in grayscale.
colorbar; %%% Put up a colorbar on the right,
axis('image'); %%%% Lock the image to have the same proportions
title('An axial slice of the brain, showing auditory cortex');
xlabel('Coronal slice number');
ylabel('Sagittal slice number');
结果:

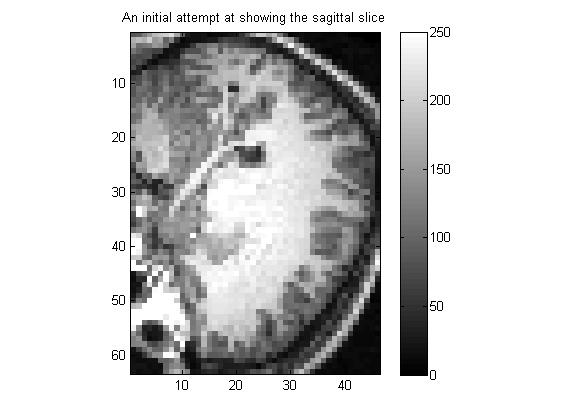
2.2 显示大脑sagittal矢状面 第20个切片的图像,以灰度图的方式显示
sagittal_slice_number = 20;
sagittal_slice_vals = subj_3danat(sagittal_slice_number,:,:);
%%%% Let's look at the variables in our workspace again
whos
%%%% Note that even though sagittal_slice_vals is just one slice,
% sagittal_slice_vals 1x63x46 23184 double array
%
% It turns out that the Matlab image command won't accept 3D matrices,
% so we need to get rid of this redundant one-voxel-wide dimension.
% Luckily, Matlab has a function that does precisely this, called squeeze.
sagittal_slice_2D = squeeze(sagittal_slice_vals);
%%% Get rid of the redundant 3rd dimension
%%%% Let's look at all the variables in our workspace
%%%% that start with "sagit"
whos sagit*
%%%% Squeeze worked! It made our sagittal slice 2D instead of 3D
%
% sagittal_slice_2D 63x46 23184 double array
%%%% Now we can plot it
figure(2);
clf;
imagesc(sagittal_slice_2D);
colormap gray; %%% Make it a grayscale plot
colorbar; %%% Show how the image intensities map onto the colormap
axis('image'); %%% Make the proportions correct
title('An initial attempt at showing the sagittal slice');
结果:
2.3 将上面大脑矢状面 图像旋转90度,以正常方式显示
caxis([ 0 250 ]); % Keep the black value at 0, but set the white value to 250
%%%% That makes the image look a lot better!
%%%% Now let's redraw the colorbar to see what the new colour-scaling is.
colorbar;
rotated_sagittal_slice = rot90(sagittal_slice_2D);
figure(3); %%% Make a new figure window, Fig.3
clf; %%% Clear the figure
imagesc(rotated_sagittal_slice);
%%% Show the image, with the brightness scaled
%%% so that the darkest is black and lightest is white
colormap gray; %%% Make it a grayscale plot
caxis([ 0 250 ]); % Keep the black value at 0, but set the white value to 250
colorbar; %%% Show how the image intensities map onto the colormap
axis('image'); %%% Make the proportions correct
title('Sagittal slice, rotated so that it is the right way up');
%%% For showing images, we often don't want to see the numbers
%%% on the x-axis and y-axis. We can turn these off using this command:
axis('off'); %%% Turn off the numbers running along the axes
结果:

2.4 利用subplot()按灰度图显示大脑冠状面9个切片的图像
figure(4); %%% Make a new figure, Fig.4
clf; %%% Clear the figure
for loop_counter = 1:9, %%% Go around 9 times, adding one to the
%%% value of loop_counter each time.
coronal_slice_number = 7*loop_counter;
%%% Loop counter goes 1,2,3,...,9
%%% so this gives slices 7,14,21,...,63
coronal_slice_vals = subj_3danat(:,coronal_slice_number,:);
%%% The coronal slice is the 2nd dimension.
%%% So, read out that slice from the 3D subj_3danat matrix
%%% The ":" signs in the 1st and 3rd coordinate positions
%%% mean "give me all the x-vals and all the z-vals in that slice"
coronal_slice_2D = squeeze(coronal_slice_vals);
%%% We have to use squeeze to take out the redundant
%%% one-voxel-wide 2nd dimension, since imagesc will
%%% not let us display a 3D matrix
rotated_coronal_slice = rot90(coronal_slice_2D);
%%% Roate the slice by 90 degrees, to make it the right way up
subplot(3,3,loop_counter);
%%% Make a 3x3 array of subplots,
%%% and draw into the "loop_counter"-th one
imagesc(rotated_coronal_slice); %%% Make the image
colormap gray; %%% Make it grayscale
caxis([ 0 250 ]);
%%% So that very intense voxels don't make everything
%%% else look dark, set the black-value to 0,
%%% and set the white value to 250, like we did above.
axis('image'); %%% Make the proportions of the image correct
axis('off'); %%% Turn off the numbers on the x- and y-axes
end; %%% The end of this for-loop
%%% If loop_counter is less than 9, then add 1 to it,
%%% and then go back to the beginning.
%%% If loop_counter is 9, then stop --- the looping is finished.
结果:

2.5 显示大脑axial 轴位 第18个切片的图像,不过以hot图的方式显示
axial_slice_number = 18;
%%%% The matrix containing a brain-full of voxels
%%%% with t-statistic numbers in them is called speech_Tmap
axial_slice_Tmap_vals = speech_Tmap(:,:,axial_slice_number);
%%%% Squeeze it, to get rid of the nuisance one-voxel wide 3rd dimension
%%%% in case there is one. (There might not always be, depending on
%%%% what type of slice you are taking. But it's a good idea to squeeze
%%%% all the time anyway, because it will make your programs more robust).
axial_slice_Tmap_2D = squeeze(axial_slice_Tmap_vals);
%%%% Now let's show this matrix as an image, in the way we did above.
figure(5);
clf;
imagesc(axial_slice_Tmap_2D);
axis('image'); %%% Make the proportions of the image correct
title('Raw unthresholded t-map');
%%%% For functional maps, a nice colormap to use is called "hot"
%%%% This makes the lowest numbers black, and then the colours
%%%% get "warmer" as the numbers increase: red, orange, yellow, white
%%%% To see a number of useful colormaps, type
%%%% help graph3d
%%%% into the Matlab command window.
colormap hot;
%%% Let's make a colorbar to show us which t-map values get shown
%%% as which colours.
colorbar;
结果:

2.6 设定一个阈值,如果矩阵中有元素大于Tmap_threshold ,就设为1,否则置为0。
Tmap_threshold = 3.5;
%%%% In Matlab, the way to figure out which voxels have values
%%%% above this threshold is to make a matrix that has a 1 in every
%%%% above-threshold voxel, and a zero everywhere else.
%%%%
%%%% In other words, we want a 1 in every voxel where the statement
%%%% "this voxel's intensity is greater than the threshold" is true.
%%%%
%%%% Here's how we do that in Matlab:
True_or_false_map = ( axial_slice_Tmap_2D > Tmap_threshold );
%%%% The matrix will have a 1 in it in every voxel where the
%%%% bracketed expression ( axial_slice_Tmap_2D > Tmap_threshold )
%%%% is true, i.e. in all the above-threshold voxels,
%%%% and a zero in all the voxels where it's false.
%%%% Making true-or-false matrices like this turns out
%%%% to be useful in all kinds of circumstances.
%%%% Let's have a look at our True_or_false_map:
figure(6);
clf;
imagesc(True_or_false_map);
axis('image');
colormap hot;
colorbar;
title('1 if t-map is above-threshold, 0 if it is below-threshold');
结果:

2.7 将阈值图矩阵和真实的图像做“点乘运算”,过滤非激活元素
Above_threshold_Tmap = True_or_false_map .* axial_slice_Tmap_2D;
%%%% Let's look at our above-treshold T-map
figure(7);
clf;
imagesc(Above_threshold_Tmap);
axis('image');
colormap hot;
colorbar;
title('The thresholded t-map');
结果:

2.8 集中对比显示,阈值化前后的效果
figure(8);
clf;
subplot(3,1,1); %%% Three rows of subplots, one column, draw in the first one
imagesc(axial_slice_Tmap_2D);
axis('image');
axis('off');
colormap hot;
colorbar;
title('Raw unthresholded t-map');
subplot(3,1,2); %%% The last argument is 2, meaning
%%% "Draw in the 2nd of the three subplots"
imagesc(True_or_false_map);
axis('image');
axis('off');
colormap hot;
colorbar;
title('1 if above-threshold, 0 if below');
subplot(3,1,3); %%% The last argument is 3, meaning
%%% "Draw in the 3rd of the three subplots"
imagesc(Above_threshold_Tmap);
axis('image');
axis('off');
colormap hot;
colorbar;
title('The thresholded t-map');
结果:

2.9 再做一下对比,对没有叠加阈值图的大脑图像,做对比对拉伸。然后,查看效果:就是激活区域,仍然是最亮的。
max_Tmap_value = max(max(axial_slice_Tmap_2D)); % Single biggest t-map score
%%%% Recall that the argument that we give to caxis is a two-element
%%%% vector, containing the maximum and minimum values we want
%%%% for the colorbar.
colorbar_min_and_max_vals = [ 0 max_Tmap_value ];
%%%% Now we can plot our thresholded and unthresholded t-maps
%%%% side-by-side, using exactly the same colour scaling in both.
figure(9);
clf;
subplot(2,1,1); %%% Two rows of subplots, one column, draw in the first one
imagesc(axial_slice_Tmap_2D);
axis('image');
axis('off');
colormap hot;
%%% Now set the colour scaling to what we want.
%%% Do this before making the colorbar
caxis(colorbar_min_and_max_vals);
colorbar;
title('t-map showing all above-zero values');
subplot(2,1,2); %%% The last argument is 2, meaning
%%% "Draw in the 2nd of the two subplots"
imagesc(Above_threshold_Tmap);
axis('image');
axis('off');
colormap hot;
caxis(colorbar_min_and_max_vals);
%%% This subplot will now have the same colour scaling as the first one
colorbar;
title('The thresholded t-map, with same colour-scaling as the plot above');
结果:

2.10 在spm中,可以通过滚动鼠标滚轮的方式,动态查看(按animation或者movie的方式)查看多层图像。
这里通过键盘按键的方式,进行了实现。不过,智能实现一次循环。
figure(10);
clf;
set(gcf,'doublebuffer','on');
for sagittal_slice_number = 1:53,
%%% There are 53 sagittal slices, and this loop
%%% goes around 53 times, adding one to the
%%% value of sagittal_slice_number each time.
sagittal_slice_vals = subj_3danat(sagittal_slice_number,:,:);
sagittal_slice_2D = squeeze(sagittal_slice_vals);
rotated_sagittal_slice = rot90(sagittal_slice_2D);
imagesc(rotated_sagittal_slice);
colormap gray;
caxis([ 0 250 ]);
axis('image');
xlabel(['Sagittal slice number ' num2str(sagittal_slice_number) ]);
title('Press any key to show the next slice');
pause;
%%% After the key press, the program continues,
end; %%% The end of this for-loop
结果:

the end 完整代码
%%%% Tutorial on how to show brain images in Matlab
%%%% Written by Rajeev Raizada, July 23, 2002.
%%%%
%%%% To run it, you need the file containing the brain images:
%%%% speech_brain_images.mat (2.3Mb)
%%%% Probably the best way to look at this program is to read through it
%%%% line by line, and paste each line into the Matlab command window
%%%% in turn. That way, you can see what effect each individual command has.
%%%%
%%%% Anything with a % sign in front of it is a comment.
%%%% These will probably show up red in your Matlab Editor.
%%%% Everything else is a Matlab command, that you can copy and
%%%% paste into the Matlab command window.
%%%%
%%%% Alternatively, you can run the program directly by typing
%%%%
%%%% showing_brain_images_tutorial
%%%%
%%%% into your Matlab command window.
%%%% Do not type ".m" at the end.
%%%% If you run the program all at once, all the Figure windows
%%%% will get made at once and will be sitting on top of each other.
%%%% You can move them around to see the ones that are hidden beneath.
%%%%
%%%% Please mail any comments or suggestions to: raj@nmr.mgh.harvard.edu
%%%%%%% First, clear the Matlab workspace of any variables
%%%%%%% that it might have stored in it, and close any existing figures
clear all; %%% Empty the workspace of all variables
close all; %%% Close any figures that might be open
%%%%%%% Load in the file containing the brain images
load speech_brain_images.mat
%%%%%%% Let's look at the variables that we've just loaded in
%%%%%%% The matlab command to do this is "whos"
%%%%%%% It shows what the variables are called, what size they are,
%%%%%%% how much memory they take up, and what data-types they are.
whos
%%%% This is the output that you should get:
%
% Name Size Bytes Class
%
% speech_Tmap 53x63x46 1228752 double array
% subj_3danat 53x63x46 1228752 double array
% This means that the variables "speech_Tmap" and "subj_3danat"
% are both 3-dimensional matrices.
% Each one has 53 rows, 63 columns, and 46 slices in the third dimension.
% Each matrix is a brain-full of voxel values.
% These particular images have already been preprocessed by SPM,
% which means that they have already been squashed and stretched
% in order to match onto a standard brain-template.
% This process of squashing and stretching is called spatial
% normalisation. The advantage of normalising our images
% onto this standard space is that we can overlap one image
% on top of another one, and they will line-up correctly.
% This is useful both for overlaying functional maps on top
% of anatomical images, and also for averaging together
% functional maps from different subjects.
%
% There are a variety of different ways of doing spatial normalisation.
% The most common one is to squash and stretch the brain images
% to fit a standard brain that was mapped out by Talairach.
% That's called putting the brain into Talairach-space, or "Talairach-ing".
% SPM uses MNI-space, which is similar to Talairach space.
% Freesurfer/FS-FAST uses a different approach: the grey
% matter is segmented, inflated, and then normalised onto
% a spherical surface.
%
% The ordering of the dimensions in our images is as follows:
%
% The first dimension is the MRI x-direction: ear-to-ear
% x specifies the x-th sagittal slice, and as you
% increase x you move from one ear to the other ear.
% (Whether this means going from right-to-left or left-to-right
% depends on whether you are using neurological or radiological convention).
% Neurological convention: left-is-left.
% Radiological convention: left-is-right.
%
% The second dimension is the MRI y-direction: back-to-front
% y specifies the y-th coronal slice, and as you
% increase y you move from the back of the brain to the front.
%
% The third dimension is the MRI z-direction: feet-to-head
% z specifies the z-th axial slice, and as you
% increase y you move from the bottom of the brain to the top.
%%%% Let's look at the 18th axial slice.
%%%% This goes through Heschl's gyrus, which is auditory cortex
axial_slice_number = 18;
%%%% The axial slices are specified in the z-direction,
%%%% which means that we want to specify the third coordinate.
%%%% We want all the data in that given z-slice,
%%%% i.e. the data for all the x-values and all the y-values
%%%% In Matlab, you say that you want all the values in
%%%% a particular dimension by putting a colon sign there.
%%%% Let's extract the data from the anatomical brain image,
%%%% which is stored in the matrix called subj_3danat
%%%% So, to extract the particular axial slice that we want,
%%%% and to get all the x-values and y-values in that slice,
%%%% we do this:
axial_slice_vals = subj_3danat(:,:,axial_slice_number);
%%%% Let's look at this matrix in a figure
figure(1); %%% Open a new figure window, Figure 1
clf; %%% Clear the figure
imagesc(axial_slice_vals);
%%% Display the matrix as an image.
%%% The columns in the matrix get displayed in the x-direction
%%% going from left to right, and the rows get displayed in
%%% the y-direction, going from top to bottom.
%%% Each pixel in the image shows the value of one entry
%%% in the matrix.
%%% The colour of each pixel is determined by the value of the
%%% entry in the matrix. Typically we want low numbers to give
%%% dark colours, and high numbers to give bright colours.
%%% Matlab has various built in colormaps that do this.
%%% For anatomical images, we typically want to view them in grayscale
%%% We achieve this with the following Matlab command
colormap gray; %%% Show the image in grayscale.
%%%% The default colormap is called "jet". It goes from
%%%% dark blue, up through light blue, yellow and red.
%%%% That's probably how your brain picture looked before
%%%% you pasted the command "colormap gray" into the command window.
%%%% Note that the command we used to show the image was not
%%%% image(), it was imagesc(). Note the "sc" after image.
%%%% This means "image scaled". The scaling means that Matlab
%%%% automatically sets it so that the lowest number in the matrix
%%%% gets shown as the first colour in the colormap, which is black
%%%% in this case, and the highest number gets shown as the
%%%% last colour in the colormap, which is white for colormap gray.
%%%% So, the darkest pixel is black and the lightest is white.
%%%% It would be nice to see how the numbers in the image
%%%% correspond to the colours in the colormap.
%%%% The Matlab command "colorbar" does this.
colorbar; %%% Put up a colorbar on the right,
%%% showing how the numbers get mapped to colours
%%%% As it happens, the axial slice is more or less the same
%%%% shape as the default Matlab figure window.
%%%% So, its proportions probably look about right.
%%%% But actually the proportions are just following the shape
%%%% of the figure window. Try grabbing a corner of the
%%%% figure window with the mouse and dragging to resize the window.
%%%% You'll see that the image gets stretched and squashed
%%%% to follow the window size.
%%%% It would be good to lock the image's aspect ratio
%%%% so that it has the correct proportions.
%%%% That way the image won't be stretched or squashed.
%%%% The command axis('image') does this.
axis('image'); %%%% Lock the image to have the same proportions
%%%% as the matrix that it is displaying
%%% Let's give our figure a title, and let's label the axes
title('An axial slice of the brain, showing auditory cortex');
xlabel('Coronal slice number');
ylabel('Sagittal slice number');
%%%% Now try stretching and squashing the figure window, and
%%%% you'll see that the image always keeps the correct proportions
%%%% Now let's try looking at a sagittal slice, e.g. slice 20
sagittal_slice_number = 20;
%%%% The sagittal slices are in the MRI x-direction, which
%%%% is the first coordinate.
%%%% So, we specify the first coordinate, and put a colon sign
%%%% in the 2nd and 3rd coordinates, meaning
%%%% "give me all the y and z values for the slice with this given x-value".
sagittal_slice_vals = subj_3danat(sagittal_slice_number,:,:);
%%%% Let's look at the variables in our workspace again
whos
%%%% Note that even though sagittal_slice_vals is just one slice,
%%%% i.e. it is 2-dimensional, it is showing up in the workspace
%%%% as a 3D matrix that is 1x63x46
%%%%
%%%% I.e. Even though we have set the x-dimension to just a single
%%%% value, namely sagittal_slice, that dimension hasn't disappeared.
%%%% It's still there in the matrix, it's just one voxel wide.
%
% sagittal_slice_vals 1x63x46 23184 double array
%
% It turns out that the Matlab image command won't accept 3D matrices,
% so we need to get rid of this redundant one-voxel-wide dimension.
% Luckily, Matlab has a function that does precisely this, called squeeze.
sagittal_slice_2D = squeeze(sagittal_slice_vals);
%%% Get rid of the redundant 3rd dimension
%%%% Let's look at all the variables in our workspace
%%%% that start with "sagit"
whos sagit*
%%%% Squeeze worked! It made our sagittal slice 2D instead of 3D
%
% sagittal_slice_2D 63x46 23184 double array
%%%% Now we can plot it
figure(2); %%% Make a new figure window, Fig.2
clf; %%% Clear the figure
imagesc(sagittal_slice_2D);
%%% Show the image, with the brightness scaled
%%% so that the darkest is black and lightest is white
colormap gray; %%% Make it a grayscale plot
colorbar; %%% Show how the image intensities map onto the colormap
axis('image'); %%% Make the proportions correct
title('An initial attempt at showing the sagittal slice');
%%%% When we look at Figure 2, we can see that it doesn't look quite right
%%%% Firstly, it's the wrong way round. The eye is looking downwards!
%%%% Secondly, it's very dark, except for one very bright point, which
%%%% is a blood vessel. What has happened is that this one very intense
%%%% point got set to white, and everything else got scaled accordingly,
%%%% meaning that everything else gets shown as a dull gray.
%%%% We want to change the scaling of our colormap so that these
%%%% less intense points get shown brighter.
%%%% If you look at the colorbar in Figure 2, you can see that
%%%% the current scaling is that black is near to zero,
%%%% and white is around 700-ish.
%%%% Maybe if we set the white-point to be 250, things would look better.
%%%% We might "over-expose" the most intense voxels and push them all
%%%% into being uniform white, but we'll get better contrast in the
%%%% parts of the brain that we care about.
%%%% The command to change the colour-scaling in Matlab is "caxis".
%%%% We give it a two numbers, put into square brackets to make
%%%% them into two-entry vector.
%%%% The first number is the intensity value to set to black.
%%%% The second number is the intensity value to set to white.
caxis([ 0 250 ]); % Keep the black value at 0, but set the white value to 250
%%%% That makes the image look a lot better!
%%%% Now let's redraw the colorbar to see what the new colour-scaling is.
colorbar;
%%% Notice that the colorbar goes from 0 to 250 now, instead of 0 to 700.
%%% That has fixed one problem, but the image is still the wrong way round.
%%% The eyes are still facing down.
%%% We want to rotate the image by 90 degrees.
%%% The way we do this in Matlab is to rotate the matrix by 90 degrees,
%%% and then redraw the newly rotated matrix.
%%% The Matlab command to rotate a matrix by 90 degrees is called "rot90".
rotated_sagittal_slice = rot90(sagittal_slice_2D);
figure(3); %%% Make a new figure window, Fig.3
clf; %%% Clear the figure
imagesc(rotated_sagittal_slice);
%%% Show the image, with the brightness scaled
%%% so that the darkest is black and lightest is white
colormap gray; %%% Make it a grayscale plot
caxis([ 0 250 ]); % Keep the black value at 0, but set the white value to 250
colorbar; %%% Show how the image intensities map onto the colormap
axis('image'); %%% Make the proportions correct
title('Sagittal slice, rotated so that it is the right way up');
%%% For showing images, we often don't want to see the numbers
%%% on the x-axis and y-axis. We can turn these off using this command:
axis('off'); %%% Turn off the numbers running along the axes
%%%%%%%%%%%%%%%%% Making subplots %%%%%%%%%%%%%%%%%%
%%%% Another useful trick is to show more than one plot
%%%% in a single figure window.
%%%% The command "subplot" does this.
%%%% Subplot gets called with three arguments:
%%%% 1. How many rows of subfigures you want
%%%% 2. How many columns of subfigures you want
%%%% 3. Which subfigure to plot in.
%%%% The subfigures are numbered in turn row-by-row
%%%% Let's make a figure window showing a 3x3 array
%%%% array of subplots, each one showing a different coronal slice
%%%% The coronal slices are the y-coordinate, the 2nd dimension,
%%%% and there are 63 of them.
%%%% This uses all the techniques that we learned above.
%%%% We'll use a for-loop
%%%% If you're pasting these lines into the Matlab command window,
%%%% you need to copy and paste all the lines below at once.
figure(4); %%% Make a new figure, Fig.4
clf; %%% Clear the figure
for loop_counter = 1:9, %%% Go around 9 times, adding one to the
%%% value of loop_counter each time.
coronal_slice_number = 7*loop_counter;
%%% Loop counter goes 1,2,3,...,9
%%% so this gives slices 7,14,21,...,63
coronal_slice_vals = subj_3danat(:,coronal_slice_number,:);
%%% The coronal slice is the 2nd dimension.
%%% So, read out that slice from the 3D subj_3danat matrix
%%% The ":" signs in the 1st and 3rd coordinate positions
%%% mean "give me all the x-vals and all the z-vals in that slice"
coronal_slice_2D = squeeze(coronal_slice_vals);
%%% We have to use squeeze to take out the redundant
%%% one-voxel-wide 2nd dimension, since imagesc will
%%% not let us display a 3D matrix
rotated_coronal_slice = rot90(coronal_slice_2D);
%%% Roate the slice by 90 degrees, to make it the right way up
subplot(3,3,loop_counter);
%%% Make a 3x3 array of subplots,
%%% and draw into the "loop_counter"-th one
imagesc(rotated_coronal_slice); %%% Make the image
colormap gray; %%% Make it grayscale
caxis([ 0 250 ]);
%%% So that very intense voxels don't make everything
%%% else look dark, set the black-value to 0,
%%% and set the white value to 250, like we did above.
axis('image'); %%% Make the proportions of the image correct
axis('off'); %%% Turn off the numbers on the x- and y-axes
end; %%% The end of this for-loop
%%% If loop_counter is less than 9, then add 1 to it,
%%% and then go back to the beginning.
%%% If loop_counter is 9, then stop --- the looping is finished.
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%
%%%% Ok, that's enough of the anatomical images.
%%%% Let's try looking at some functional maps.
%%%% A functional map is an image, just like an anatomical,
%%%% except that each voxel's intensity represents the statistical
%%%% significance of BOLD activiation at that point, rather than
%%%% showing what type of anatomical tissue is there.
%%%%
%%%% There are various types of statistical values that
%%%% can be used in functional maps: t-values, Z-values, p-values.
%%%% A common one to use is the t-value (this is exactly the same
%%%% number as in a regular t-test).
%%%% The bigger the t-value, the more statistically significant
%%%% is the neural activation at that part of the brain.
%%%% A brain-full of t-values is called a t-map.
%%%%
%%%% Eventually we'll want to overlay these t-maps on top of
%%%% the anatomical images, but it turns out that this a little trickier.
%%%% An accompanying program overlaying_Tmaps_tutorial.m
%%%% shows how to do that.
%%%% That program is not meant to be introductory, it assumes
%%%% that you have already worked through this one first.
%%%%
%%%% Let's look at the t-map in the same axial slice that we
%%%% looked at in the anatomical image in Figure 1.
%%%% It's the 18th axial slice.
%%%% This goes through Heschl's gyrus, which is auditory cortex
axial_slice_number = 18;
%%%% The matrix containing a brain-full of voxels
%%%% with t-statistic numbers in them is called speech_Tmap
axial_slice_Tmap_vals = speech_Tmap(:,:,axial_slice_number);
%%%% Squeeze it, to get rid of the nuisance one-voxel wide 3rd dimension
%%%% in case there is one. (There might not always be, depending on
%%%% what type of slice you are taking. But it's a good idea to squeeze
%%%% all the time anyway, because it will make your programs more robust).
axial_slice_Tmap_2D = squeeze(axial_slice_Tmap_vals);
%%%% Now let's show this matrix as an image, in the way we did above.
figure(5);
clf;
imagesc(axial_slice_Tmap_2D);
axis('image'); %%% Make the proportions of the image correct
title('Raw unthresholded t-map');
%%%% For functional maps, a nice colormap to use is called "hot"
%%%% This makes the lowest numbers black, and then the colours
%%%% get "warmer" as the numbers increase: red, orange, yellow, white
%%%% To see a number of useful colormaps, type
%%%% help graph3d
%%%% into the Matlab command window.
colormap hot;
%%% Let's make a colorbar to show us which t-map values get shown
%%% as which colours.
colorbar;
%%%%%%%%%%%% How to show a thresholded t-map %%%%%%%%
%
% You can see from Figure 5 that the strongest activity is in auditory
% cortex, bilaterally. That's what we'd expect, given that it's speech.
% For some unknown reason the right cortex is activated more than the left
% in this particular image.
%
% Often we want to show a thresholded statistical map.
% To do this, we need to do two things:
%
% 1. Figure out which voxels exceed our threshold.
% 2. Make a map that shows these voxels' values, but it zero everywhere else.
%%%% First of all, we need to set a threshold
Tmap_threshold = 3.5;
%%%% In Matlab, the way to figure out which voxels have values
%%%% above this threshold is to make a matrix that has a 1 in every
%%%% above-threshold voxel, and a zero everywhere else.
%%%%
%%%% In other words, we want a 1 in every voxel where the statement
%%%% "this voxel's intensity is greater than the threshold" is true.
%%%%
%%%% Here's how we do that in Matlab:
True_or_false_map = ( axial_slice_Tmap_2D > Tmap_threshold );
%%%% The matrix will have a 1 in it in every voxel where the
%%%% bracketed expression ( axial_slice_Tmap_2D > Tmap_threshold )
%%%% is true, i.e. in all the above-threshold voxels,
%%%% and a zero in all the voxels where it's false.
%%%% Making true-or-false matrices like this turns out
%%%% to be useful in all kinds of circumstances.
%%%% Let's have a look at our True_or_false_map:
figure(6);
clf;
imagesc(True_or_false_map);
axis('image');
colormap hot;
colorbar;
title('1 if t-map is above-threshold, 0 if it is below-threshold');
%%%% Notice that everything is black and white,
%%%% even though we are using the colormap hot.
%%%% That's because everything is either 0 or 1,
%%%% so we're not seeing any of the nice mid-value oranges and yellows.
%%%%
%%%% So, what we want is to combine the True_or_false_map
%%%% with the map of continuous-valued actual t-map numbers.
%%%%
%%%% The way to do this is to multiply the True_or_false_map
%%%% element-by-element with the actual t-map.
%%%% "Element-by-element" means that, say, the Row-3,Column-5 entry
%%%% in the True_or_false_map gets multiplied by the Row-3,Column-5 entry
%%%% in the actual t-map.
%%%% Note that this is NOT the same as matrix multiplication!
%%%% Also, the two matrices must be exactly the same size as each other.
%%%%
%%%% In above-threshold voxels, the result of the multiplication will be:
%%%%
%%%% 1 * t-map-value
%%%%
%%%% ( It's multiplied by one because the
%%%% True_or_false_map is 1 here, because this voxel is above-threshold )
%%%%
%%%% In below-threshold voxels, the result of the multiplication will be:
%%%%
%%%% 0 * t-map-value because these voxels are below threshold
%%%%
%%%% The way to do element-by-element multiplication in Matlab is
%%%% to put a dot in front of the * sign.
%%%% So, A*B means "A matrix-multiplied by B",
%%%% and A.*B (note the dot) means "A element-by-element multiplied by B"
Above_threshold_Tmap = True_or_false_map .* axial_slice_Tmap_2D;
%%%% Let's look at our above-treshold T-map
figure(7);
clf;
imagesc(Above_threshold_Tmap);
axis('image');
colormap hot;
colorbar;
title('The thresholded t-map');
%%%% It's interesting to look at the three maps we made side by side.
%%%% We'll use the subplot command from above, with one column and three rows.
figure(8);
clf;
subplot(3,1,1); %%% Three rows of subplots, one column, draw in the first one
imagesc(axial_slice_Tmap_2D);
axis('image');
axis('off');
colormap hot;
colorbar;
title('Raw unthresholded t-map');
subplot(3,1,2); %%% The last argument is 2, meaning
%%% "Draw in the 2nd of the three subplots"
imagesc(True_or_false_map);
axis('image');
axis('off');
colormap hot;
colorbar;
title('1 if above-threshold, 0 if below');
subplot(3,1,3); %%% The last argument is 3, meaning
%%% "Draw in the 3rd of the three subplots"
imagesc(Above_threshold_Tmap);
axis('image');
axis('off');
colormap hot;
colorbar;
title('The thresholded t-map');
%%%%% If you look carefully at Figure 8, you'll notice that the colours
%%%%% in the above-threshold voxels in the uppermost plot are not
%%%%% exactly the same as the colours in the lowermost plot.
%%%%% That's because the colorbar scalings are slightly different.
%%%%%
%%%%% The top image, which is of the raw untresholded map,
%%%%% has numbers less than -2 in it, and these are shown as black.
%%%%% Notice how the top colorbar goes from -2something to 8something.
%%%%%
%%%%% But the lowermost image, of the thresholded map,
%%%%% doesn't have any numbers in it less than zero.
%%%%% But it still goes just as high, as you can see from the colobar.
%%%%% So, the oranges and yellows get spread out along the number-line
%%%%% a little differently.
%%%%%
%%%%% When you are presenting images side-by-side, it's often better
%%%%% to have the same colour-scaling for all of them.
%%%%% Otherwise it can be misleading.
%%%%% We learned above how to specify a fixed colobar scaling
%%%%% by using the command "caxis". Let's do that for our side-by-side plots.
%%%%%
%%%%% A good setting for the colorbar scaling would be to have zero shown
%%%%% as black, and to have the maximum value in the map shown as white.
%%%%% Note that setting zero to be the black-point will look the same as
%%%%% if we had thresholded the t-map at zero, because we won't see
%%%%% any of the negative parts of the t-map any more.
%%%%%
%%%%% To find what the maximum value is, we can use the Matlab command "max".
%%%%% It turns out that if we apply "max" to a 2D-matrix, it gives us
%%%%% a row-vector of numbers giving the maximum value for each column.
%%%%% But we just want the maximum value in the whole matrix.
%%%%% So, we use max twice. That way, we get a single number that is
%%%%% the maximum in the entire matrix.
max_Tmap_value = max(max(axial_slice_Tmap_2D)); % Single biggest t-map score
%%%% Recall that the argument that we give to caxis is a two-element
%%%% vector, containing the maximum and minimum values we want
%%%% for the colorbar.
colorbar_min_and_max_vals = [ 0 max_Tmap_value ];
%%%% Now we can plot our thresholded and unthresholded t-maps
%%%% side-by-side, using exactly the same colour scaling in both.
figure(9);
clf;
subplot(2,1,1); %%% Two rows of subplots, one column, draw in the first one
imagesc(axial_slice_Tmap_2D);
axis('image');
axis('off');
colormap hot;
%%% Now set the colour scaling to what we want.
%%% Do this before making the colorbar
caxis(colorbar_min_and_max_vals);
colorbar;
title('t-map showing all above-zero values');
subplot(2,1,2); %%% The last argument is 2, meaning
%%% "Draw in the 2nd of the two subplots"
imagesc(Above_threshold_Tmap);
axis('image');
axis('off');
colormap hot;
caxis(colorbar_min_and_max_vals);
%%% This subplot will now have the same colour scaling as the first one
colorbar;
title('The thresholded t-map, with same colour-scaling as the plot above');
%%%%%%%% Doing a "fly-through" of the slices %%%%%%%%%%%%
%
% To make an animation of a "fly through" the slices,
% we use a loop very similar to the one we used for making
% Figure 4, with its nine subplots of various slices.
% Except this time, instead of showing the different slices
% next to each other, we'll show them all in the same place
% but in quick sequence, giving the effect of a movie.
figure(10); %%% Make a new figure
clf; %%% Clear the figure
%%% It turns out that there's a nifty Matlab trick that allows us
%%% to rapidly redraw new images, without the window flickering.
%%% The command below does this. Preventing the flicker makes
%%% the movie look a *lot* nicer.
set(gcf,'doublebuffer','on');
% gcf means "get current figure".
% So, this means "set double-buffering to be on in the current figure".
% Double-buffering means that Matlab draws the figure into a virtual
% figure window in memory first, before copying it to the actual screen.
% That's not really important, the main thing is that it stops flicker.
for sagittal_slice_number = 1:53,
%%% There are 53 sagittal slices, and this loop
%%% goes around 53 times, adding one to the
%%% value of sagittal_slice_number each time.
sagittal_slice_vals = subj_3danat(sagittal_slice_number,:,:);
%%% The sagittal slice is the 1st dimension, the x-direction.
%%% So, read out that slice from the 3D subj_3danat matrix
%%% The ":" signs in the 2nd and 3rd coordinate positions
%%% mean "give me all the y-vals and all the z-vals in that slice"
sagittal_slice_2D = squeeze(sagittal_slice_vals);
%%% We have to use squeeze to take out the redundant
%%% one-voxel-wide 1st dimension, since imagesc will
%%% not let us display a 3D matrix
rotated_sagittal_slice = rot90(sagittal_slice_2D);
%%% Roate the slice by 90 degrees, to make it the right way up
imagesc(rotated_sagittal_slice); %%% Make the image
colormap gray; %%% Make it grayscale
caxis([ 0 250 ]);
%%% So that very intense voxels don't make everything
%%% else look dark, set the black-value to 0,
%%% and set the white value to 250, like we did above.
axis('image'); %%% Make the proportions of the image correct
%%%% Let's show in the x-axis label which slice we're at.
%%%% We can do this by making a compound string containing
%%%% so text, and then the slice-number (which is a numeral),
%%%% turned into a string. We have to change the number into
%%%% a string because to a computer the number 5 is one more than 4,
%%%% but the string "5" is an particular ASCII character.
%%%% The Matlab command num2str does this for us.
%%%% We put the whole compound string inside [ square brackets ]
xlabel(['Sagittal slice number ' num2str(sagittal_slice_number) ]);
%%%%% Before going to the next slice, wait for the user to press a key
title('Press any key to show the next slice');
pause; %%% This pause command waits for a key-press
%%% After the key press, the program continues,
%%% reaches the "end" command below, and that "end"
%%% either takes us back to beginning of the loop,
%%% to show the next slice, or stops at the last slice.
end; %%% The end of this for-loop
%%% If sagittal_slice_number is less than 53, then add 1 to it,
%%% and then go back to the beginning.
%%% If sagittal_slice_number is 53, then stop.