The most common useful indexes have been collected by Holliday et al (Holliday, JD., Hu, C-Y. and Willett, P. (2002) Combinatorial Chemistry and High Throughput Screening 5, 155-166) These are shown in the table, and can be referred to, by name, in applications and toolkits calls which allow user defined similarity functions.
| Measure | Range | Formula |
| Cosine | 0.0,1.0 | 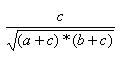 |
| Dice | 0.0,1.0 | 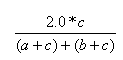 |
| Euclid | 0.0,1.0 | 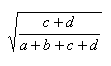 |
| Forbes | 0.0,∞ | 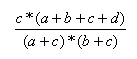 |
| Hamman | -1.0,1.0 | 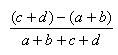 |
| Jaccard | 0.0,1.0 | |
| Kulczynski | 0.0,1.0 | 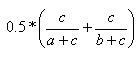 |
| Manhattan | 1.0,0.0 | 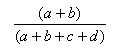 |
| Matching | 0.0,1.0 | 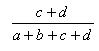 |
| Pearson | -1.0,1.0 | 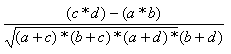 |
| Rogers-Tanimoto | 0.0,1.0 | 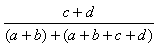 |
| Russell-Rao | 0.0,1.0 | 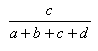 |
| Simpson | 0.0,1.0 | 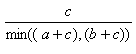 |
| Tanimoto | 0.0,1.0 | 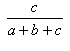 |
| Yule | -1.0,1.0 | 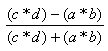 |
Notes
- The Tanimoto and Jaccard indexes are the same.
- The Forbes index has no upper limit.
- The Manhattan index is a distance = 1.0 - Matching index
- The Kulczynski index is the mean of the individual substructure similarities
- The Simpson index is the best of the individual substructure similarities
- The Dice index is the ratio of the bits in common to the arithmetic mean of the number of on bits in the two items.
- The Cosine index is the ration of the bits in common to the geometric mean of the number of on bits in the two items.
from : http://www.daylight.com/dayhtml/doc/theory/theory.finger.html