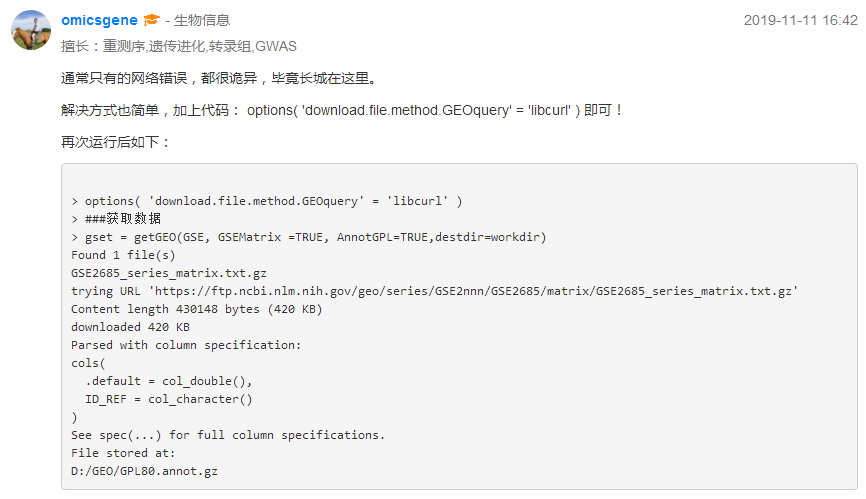
其实加载GEOquery包的时候就出现options()的这行代码
> library(GEOquery) 载入需要的程辑包:Biobase 载入需要的程辑包:BiocGenerics 载入需要的程辑包:parallel 载入程辑包:‘BiocGenerics’ The following objects are masked from ‘package:parallel’: clusterApply, clusterApplyLB, clusterCall, clusterEvalQ, clusterExport, clusterMap, parApply, parCapply, parLapply, parLapplyLB, parRapply, parSapply, parSapplyLB The following objects are masked from ‘package:stats’: IQR, mad, sd, var, xtabs The following objects are masked from ‘package:base’: anyDuplicated, append, as.data.frame, basename, cbind, colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget, order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank, rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply, union, unique, unsplit, which, which.max, which.min Welcome to Bioconductor Vignettes contain introductory material; view with 'browseVignettes()'. To cite Bioconductor, see 'citation("Biobase")', and for packages 'citation("pkgname")'. Setting options('download.file.method.GEOquery'='auto') #### Setting options('GEOquery.inmemory.gpl'=FALSE)
其实不仅仅是在加载Bioconductor包的时候会有这样的连不上的报错,在下载有些数据的时候也会,比如下载GEO的数据:
> library(GEOquery) Setting options('download.file.method.GEOquery'='auto') Setting options('GEOquery.inmemory.gpl'=FALSE) > if(!file.exists(f)){} NULL > if(!file.exists(f)){ + gset <- getGEO("GSE42872",destdir = ".",AnnotGPL = F,getGPL = F) + save(gset,file = f) + } Found 1 file(s) GSE42872_series_matrix.txt.gz 试开URL’https://ftp.ncbi.nlm.nih.gov/geo/series/GSE42nnn/GSE42872/matrix/GSE42872_series_matrix.txt.gz' Error in download.file(sprintf("https://ftp.ncbi.nlm.nih.gov/geo/series/%s/%s/matrix/%s", : 无法打开URL'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE42nnn/GSE42872/matrix/GSE42872_series_matrix.txt.gz' 3. download.file(sprintf("https://ftp.ncbi.nlm.nih.gov/geo/series/%s/%s/matrix/%s", stub, GEO, b[i]), destfile = destfile, mode = "wb", method = getOption("download.file.method.GEOquery")) 2. getAndParseGSEMatrices(GEO, destdir, AnnotGPL = AnnotGPL, getGPL = getGPL, parseCharacteristics = parseCharacteristics) 1. getGEO("GSE42872", destdir = ".", AnnotGPL = F, getGPL = F)
当把options('download.file.method.GEOquery'='auto') 改成 options('download.file.method.GEOquery'='libcurl')的时候就可以下载了!、
> options('download.file.method.GEOquery'='libcurl') > if(!file.exists(f)){ + gset <- getGEO("GSE42872",destdir = ".",AnnotGPL = F,getGPL = F) + save(gset,file = f) + } Found 1 file(s) GSE42872_series_matrix.txt.gz 试开URL’https://ftp.ncbi.nlm.nih.gov/geo/series/GSE42nnn/GSE42872/matrix/GSE42872_series_matrix.txt.gz' Content length 768865 bytes (750 KB) downloaded 750 KB Parsed with column specification: cols( ID_REF = col_double(), GSM1052615 = col_double(), GSM1052616 = col_double(), GSM1052617 = col_double(), GSM1052618 = col_double(), GSM1052619 = col_double(), GSM1052620 = col_double() ) |===========================================================| 100% 1 MB > class(gset) [1] "list" > length(gset) [1] 1 > gset $GSE42872_series_matrix.txt.gz ExpressionSet (storageMode: lockedEnvironment) assayData: 33297 features, 6 samples element names: exprs protocolData: none phenoData sampleNames: GSM1052615 GSM1052616 ... GSM1052620 (6 total) varLabels: title geo_accession ... cell type:ch1 (34 total) varMetadata: labelDescription featureData: none experimentData: use 'experimentData(object)' pubMedIds: 24469106 Annotation: GPL6244
更正一下:有的时候也不定有用,咳咳。。。比如gpl6244 <- getGEO("GPL6244",destdir = "."),下载个平台注释文件老是下部完整就完了,无奈!