DNA sequence
Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Total Submission(s): 2427 Accepted Submission(s): 1198
Problem Description
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
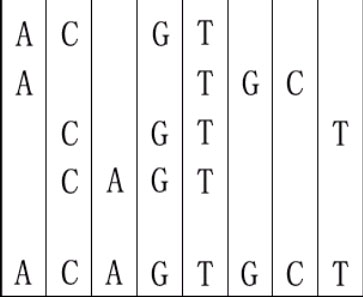
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.

Input
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
Output
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
Sample Input
1
4
ACGT
ATGC
CGTT
CAGT
Sample Output
8
Author
LL
对公共串的每一位进行搜索,并使用IDA*剪枝
1 //2017-03-07 2 #include <iostream> 3 #include <cstdio> 4 #include <cstring> 5 6 using namespace std; 7 8 string DNA[10]; 9 char ch[4] = {'A', 'T', 'C', 'G'}; 10 int deep, n, pos[10];//pos[i]表示第i个DNA串匹配到了pos[i]位 11 bool ok; 12 13 int Astar()//估价函数,返回当前状态最少还需要多少位 14 { 15 int h = 0; 16 for(int i = 0; i < n; i++) 17 if(h < DNA[i].length()-pos[i]) 18 h = DNA[i].length()-pos[i]; 19 return h; 20 } 21 22 void IDAstar(int step)//枚举所求串每一位的字符,step表示所求串当前处理到哪一位。 23 { 24 if(ok)return; 25 int h = Astar(); 26 if(step + h > deep)return;//剪枝 27 if(h == 0){//h为0表示短串全部处理完 28 ok = true; 29 return; 30 } 31 for(int i = 0; i < 4; i++) 32 { 33 int tmp[10]; 34 for(int j = 0; j < n; j++)tmp[j] = pos[j]; 35 bool fg = false; 36 for(int j = 0; j < n; j++) 37 { 38 if(DNA[j][pos[j]] == ch[i]){ 39 fg = true;//表示所求串第step为可以是ch[i] 40 pos[j]++; 41 } 42 } 43 if(fg){ 44 IDAstar(step+1); 45 } 46 for(int j = 0; j < n; j++)pos[j] = tmp[j]; 47 } 48 } 49 50 int main() 51 { 52 int T; 53 cin>>T; 54 while(T--) 55 { 56 cin>>n; 57 deep = 0; 58 for(int i = 0; i < n; i++) 59 { 60 cin>>DNA[i]; 61 if(DNA[i].length() > deep)deep = DNA[i].length(); 62 } 63 ok = false; 64 while(!ok) 65 { 66 memset(pos, 0, sizeof(pos)); 67 IDAstar(0); 68 deep++; 69 } 70 cout<<deep-1<<endl; 71 } 72 73 return 0; 74 }