https://www.rdocumentation.org/packages/DESeq2/versions/1.12.3/topics/estimateSizeFactorsForMatrix
1.描述
Given a matrix or data frame of count data, this function estimates the size factors as follows: Each column is divided by the geometric means of the rows.
The median of these ratios (skipping the genes with a geometric mean of zero) is used as the size factor for this column.
每列被每行的均值除?
一般不会直接调用它,而是通过调用estimateSizeFactors来间接调用它。
2.论文介绍
https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-10-r106
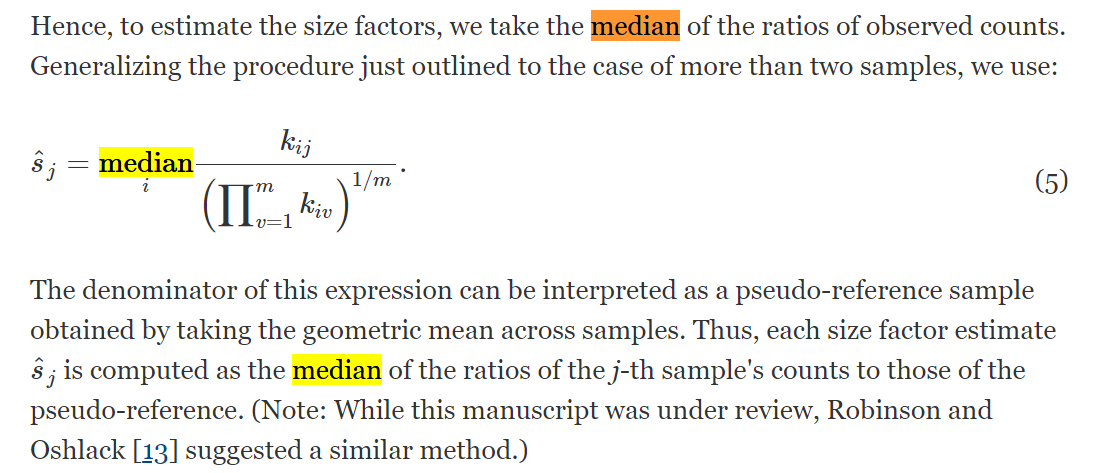
上式中,i是gene,j是cell,k矩阵是n*m的,分母相当于gene_i的所在行的均值。结合函数的描述,计算过程:每列/行的均值,在每列中选择除了0之外的数的中位数,作为每个cell的size因子。这就是比例中位数方法。
3.源码
function (counts, locfunc = stats::median, geoMeans, controlGenes, type = c("ratio", "poscounts")) { type <- match.arg(type, c("ratio", "poscounts"))#"ratio",返回的是ratio if (missing(geoMeans)) {#默认进入这条判断 incomingGeoMeans <- FALSE if (type == "ratio") { loggeomeans <- rowMeans(log(counts))#计算log后行的均值 } else if (type == "poscounts") { lc <- log(counts) lc[!is.finite(lc)] <- 0 loggeomeans <- rowMeans(lc) allZero <- rowSums(counts) == 0 loggeomeans[allZero] <- -Inf } } else { incomingGeoMeans <- TRUE if (length(geoMeans) != nrow(counts)) { stop("geoMeans should be as long as the number of rows of counts") } loggeomeans <- log(geoMeans) } if (all(is.infinite(loggeomeans))) {#有0值很正常,到了这里就退出??? stop("every gene contains at least one zero, cannot compute log geometric means") } sf <- if (missing(controlGenes)) { apply(counts, 2, function(cnts) { exp(locfunc((log(cnts) - loggeomeans)[is.finite(loggeomeans) & cnts > 0])) }) } else { if (!(is.numeric(controlGenes) | is.logical(controlGenes))) { stop("controlGenes should be either a numeric or logical vector") } loggeomeansSub <- loggeomeans[controlGenes] apply(counts[controlGenes, , drop = FALSE], 2, function(cnts) { exp(locfunc((log(cnts) - loggeomeansSub)[is.finite(loggeomeansSub) & cnts > 0])) }) } if (incomingGeoMeans) { sf <- sf/exp(mean(log(sf))) } sf }
但是源码没有跑通。